Mol:FL1CBANC0002
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 46 0 0 0 0 0 0 0 0999 V2000 | + | 43 46 0 0 0 0 0 0 0 0999 V2000 |
| − | 1.8476 0.2370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8476 0.2370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5174 0.6237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5174 0.6237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5174 1.3971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5174 1.3971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8476 1.7838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8476 1.7838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1778 1.3971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1778 1.3971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1778 0.6237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1778 0.6237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5012 1.7878 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5012 1.7878 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0215 1.4860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0215 1.4860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6191 1.8310 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6191 1.8310 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1995 1.4958 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1995 1.4958 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8473 1.8698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8473 1.8698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4951 1.4958 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4951 1.4958 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4951 0.7478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4951 0.7478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8473 0.3738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8473 0.3738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1995 0.7478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1995 0.7478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9760 0.4702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9760 0.4702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5012 2.2502 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5012 2.2502 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6451 0.3161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6451 0.3161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0649 0.3077 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0649 0.3077 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6266 -0.7530 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6266 -0.7530 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3083 -1.1465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3083 -1.1465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8352 -0.8424 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.8352 -0.8424 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.3521 -1.1408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3521 -1.1408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3521 -1.8804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3521 -1.8804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9926 -2.2502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9926 -2.2502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6331 -1.8804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6331 -1.8804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6331 -1.1408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6331 -1.1408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9926 -0.7711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9926 -0.7711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1014 -2.1508 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1014 -2.1508 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0599 -1.0802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0599 -1.0802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5095 -0.7515 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -0.5095 -0.7515 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -1.1461 -1.1190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1461 -1.1190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8287 -0.7250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8287 -0.7250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4245 -1.0690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4245 -1.0690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0336 -0.7172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0336 -0.7172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6428 -1.0690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6428 -1.0690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6428 -1.7724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6428 -1.7724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0336 -2.1241 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0336 -2.1241 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4245 -1.7724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4245 -1.7724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1014 -2.0372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1014 -2.0372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5095 -0.2576 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5095 -0.2576 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5049 2.0202 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5049 2.0202 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4459 2.3586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4459 2.3586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 5 7 1 0 0 0 0 | + | 5 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 11 2 0 0 0 0 | + | 10 11 2 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 10 1 0 0 0 0 | + | 15 10 1 0 0 0 0 |
| − | 13 16 1 0 0 0 0 | + | 13 16 1 0 0 0 0 |
| − | 7 17 2 0 0 0 0 | + | 7 17 2 0 0 0 0 |
| − | 6 18 1 0 0 0 0 | + | 6 18 1 0 0 0 0 |
| − | 2 19 1 0 0 0 0 | + | 2 19 1 0 0 0 0 |
| − | 20 21 2 0 0 0 0 | + | 20 21 2 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 2 0 0 0 0 | + | 23 24 2 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 2 0 0 0 0 | + | 25 26 2 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 2 0 0 0 0 | + | 27 28 2 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 26 29 1 0 0 0 0 | + | 26 29 1 0 0 0 0 |
| − | 20 30 1 0 0 0 0 | + | 20 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 2 0 0 0 0 | + | 34 35 2 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 39 34 1 0 0 0 0 | + | 39 34 1 0 0 0 0 |
| − | 37 40 1 0 0 0 0 | + | 37 40 1 0 0 0 0 |
| − | 31 41 1 0 0 0 0 | + | 31 41 1 0 0 0 0 |
| − | 22 1 1 6 0 0 0 | + | 22 1 1 6 0 0 0 |
| − | 4 42 1 0 0 0 0 | + | 4 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 42 43 | + | M SAL 1 2 42 43 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 45 2.5049 2.0202 | + | M SVB 1 45 2.5049 2.0202 |
| − | S SKP 8 | + | S SKP 8 |
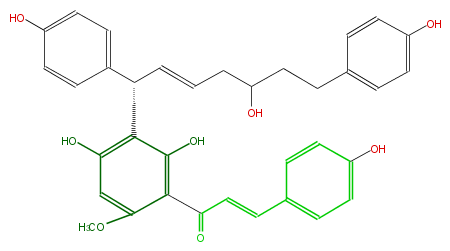
| − | ID FL1CBANC0002 | + | ID FL1CBANC0002 |
| − | KNApSAcK_ID C00008124 | + | KNApSAcK_ID C00008124 |
| − | NAME Calyxin B | + | NAME Calyxin B |
| − | CAS_RN 164991-53-1 | + | CAS_RN 164991-53-1 |
| − | FORMULA C35H34O8 | + | FORMULA C35H34O8 |
| − | EXACTMASS 582.225368064 | + | EXACTMASS 582.225368064 |
| − | AVERAGEMASS 582.63966 | + | AVERAGEMASS 582.63966 |
| − | SMILES Oc(c2[C@H](c(c4)ccc(O)c4)C=CCC(O)CCc(c3)ccc(O)c3)c(c(OC)cc2O)C(=O)C=Cc(c1)ccc(c1)O | + | SMILES Oc(c2[C@H](c(c4)ccc(O)c4)C=CCC(O)CCc(c3)ccc(O)c3)c(c(OC)cc2O)C(=O)C=Cc(c1)ccc(c1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 46 0 0 0 0 0 0 0 0999 V2000
1.8476 0.2370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5174 0.6237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5174 1.3971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8476 1.7838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1778 1.3971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1778 0.6237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5012 1.7878 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0215 1.4860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6191 1.8310 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1995 1.4958 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8473 1.8698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4951 1.4958 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4951 0.7478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8473 0.3738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1995 0.7478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9760 0.4702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5012 2.2502 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6451 0.3161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0649 0.3077 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6266 -0.7530 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3083 -1.1465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8352 -0.8424 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.3521 -1.1408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3521 -1.8804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9926 -2.2502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6331 -1.8804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6331 -1.1408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9926 -0.7711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1014 -2.1508 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0599 -1.0802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5095 -0.7515 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-1.1461 -1.1190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8287 -0.7250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4245 -1.0690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0336 -0.7172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6428 -1.0690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6428 -1.7724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0336 -2.1241 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4245 -1.7724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1014 -2.0372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5095 -0.2576 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5049 2.0202 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4459 2.3586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
5 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 11 2 0 0 0 0
11 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 10 1 0 0 0 0
13 16 1 0 0 0 0
7 17 2 0 0 0 0
6 18 1 0 0 0 0
2 19 1 0 0 0 0
20 21 2 0 0 0 0
21 22 1 0 0 0 0
22 23 1 0 0 0 0
23 24 2 0 0 0 0
24 25 1 0 0 0 0
25 26 2 0 0 0 0
26 27 1 0 0 0 0
27 28 2 0 0 0 0
28 23 1 0 0 0 0
26 29 1 0 0 0 0
20 30 1 0 0 0 0
30 31 1 0 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 35 2 0 0 0 0
35 36 1 0 0 0 0
36 37 2 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
39 34 1 0 0 0 0
37 40 1 0 0 0 0
31 41 1 0 0 0 0
22 1 1 6 0 0 0
4 42 1 0 0 0 0
42 43 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 42 43
M SBL 1 1 45
M SMT 1 OCH3
M SVB 1 45 2.5049 2.0202
S SKP 8
ID FL1CBANC0002
KNApSAcK_ID C00008124
NAME Calyxin B
CAS_RN 164991-53-1
FORMULA C35H34O8
EXACTMASS 582.225368064
AVERAGEMASS 582.63966
SMILES Oc(c2[C@H](c(c4)ccc(O)c4)C=CCC(O)CCc(c3)ccc(O)c3)c(c(OC)cc2O)C(=O)C=Cc(c1)ccc(c1)O
M END