Mol:FL1CAAGS0009
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 53 57 0 0 0 0 0 0 0 0999 V2000 | + | 53 57 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.3373 0.4820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3373 0.4820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3373 -0.3430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3373 -0.3430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0518 -0.7555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0518 -0.7555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7662 -0.3430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7662 -0.3430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7662 0.4820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7662 0.4820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0518 0.8945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0518 0.8945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4807 -0.7555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4807 -0.7555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1952 -0.3430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1952 -0.3430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1952 0.4820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1952 0.4820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9097 0.8945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9097 0.8945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6241 0.4820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6241 0.4820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3386 0.8945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3386 0.8945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3386 1.7195 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3386 1.7195 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6241 2.1320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6241 2.1320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9097 1.7195 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9097 1.7195 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4807 -1.5805 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4807 -1.5805 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6331 -1.4358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6331 -1.4358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1552 -2.1086 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1552 -2.1086 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3800 -1.8254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3800 -1.8254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4312 -1.9769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4312 -1.9769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0466 -1.3039 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0466 -1.3039 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8218 -1.5870 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8218 -1.5870 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0331 -1.0185 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0331 -1.0185 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0688 -1.9622 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0688 -1.9622 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6262 -1.8928 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6262 -1.8928 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4907 -2.4787 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4907 -2.4787 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0518 -1.4667 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0518 -1.4667 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3494 0.8785 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3494 0.8785 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6163 -0.7038 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6163 -0.7038 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3375 0.8118 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3375 0.8118 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.0198 2.1128 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.0198 2.1128 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8306 1.0078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8306 1.0078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4180 0.2931 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4180 0.2931 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6197 0.5023 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6197 0.5023 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8262 0.2754 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8262 0.2754 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2387 0.9902 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2387 0.9902 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0371 0.7811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0371 0.7811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1941 1.3669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1941 1.3669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6740 1.6440 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6740 1.6440 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3138 0.5246 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3138 0.5246 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8667 0.5522 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8667 0.5522 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7911 -0.1377 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7911 -0.1377 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.5366 1.8425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.5366 1.8425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1240 1.1278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1240 1.1278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3257 1.3370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3257 1.3370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5322 1.1101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5322 1.1101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9447 1.8249 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9447 1.8249 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7431 1.6158 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7431 1.6158 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9001 2.2016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9001 2.2016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3800 2.4787 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3800 2.4787 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.0198 1.3593 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.0198 1.3593 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.5727 1.3869 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.5727 1.3869 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4971 0.6970 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4971 0.6970 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 11 2 0 0 0 0 | + | 10 11 2 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 10 1 0 0 0 0 | + | 15 10 1 0 0 0 0 |
| − | 7 16 2 0 0 0 0 | + | 7 16 2 0 0 0 0 |
| − | 17 18 1 1 0 0 0 | + | 17 18 1 1 0 0 0 |
| − | 18 19 1 1 0 0 0 | + | 18 19 1 1 0 0 0 |
| − | 20 19 1 1 0 0 0 | + | 20 19 1 1 0 0 0 |
| − | 20 21 1 0 0 0 0 | + | 20 21 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 17 1 0 0 0 0 | + | 22 17 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 17 24 1 0 0 0 0 | + | 17 24 1 0 0 0 0 |
| − | 18 25 1 0 0 0 0 | + | 18 25 1 0 0 0 0 |
| − | 19 26 1 0 0 0 0 | + | 19 26 1 0 0 0 0 |
| − | 3 27 1 0 0 0 0 | + | 3 27 1 0 0 0 0 |
| − | 1 28 1 0 0 0 0 | + | 1 28 1 0 0 0 0 |
| − | 29 23 1 0 0 0 0 | + | 29 23 1 0 0 0 0 |
| − | 5 30 1 0 0 0 0 | + | 5 30 1 0 0 0 0 |
| − | 27 20 1 0 0 0 0 | + | 27 20 1 0 0 0 0 |
| − | 13 31 1 0 0 0 0 | + | 13 31 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 32 40 1 0 0 0 0 | + | 32 40 1 0 0 0 0 |
| − | 33 41 1 0 0 0 0 | + | 33 41 1 0 0 0 0 |
| − | 34 42 1 0 0 0 0 | + | 34 42 1 0 0 0 0 |
| − | 43 44 1 1 0 0 0 | + | 43 44 1 1 0 0 0 |
| − | 44 45 1 1 0 0 0 | + | 44 45 1 1 0 0 0 |
| − | 46 45 1 1 0 0 0 | + | 46 45 1 1 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 48 43 1 0 0 0 0 | + | 48 43 1 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 49 50 1 0 0 0 0 | + | 49 50 1 0 0 0 0 |
| − | 43 51 1 0 0 0 0 | + | 43 51 1 0 0 0 0 |
| − | 44 52 1 0 0 0 0 | + | 44 52 1 0 0 0 0 |
| − | 45 53 1 0 0 0 0 | + | 45 53 1 0 0 0 0 |
| − | 46 39 1 0 0 0 0 | + | 46 39 1 0 0 0 0 |
| − | 28 35 1 0 0 0 0 | + | 28 35 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL1CAAGS0009 | + | ID FL1CAAGS0009 |
| − | KNApSAcK_ID C00014505 | + | KNApSAcK_ID C00014505 |
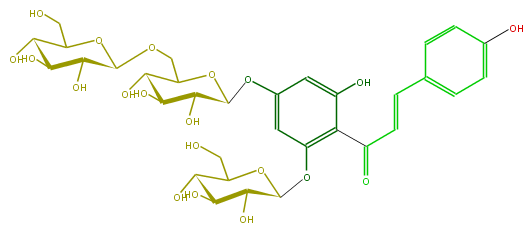
| − | NAME Chalconaringenin 2'-O-glucoside 4'-O-gentobioside;4,2',4',6'-Tetrahydroxychalcone 2'-O-glucoside 4'-O-gentobioside | + | NAME Chalconaringenin 2'-O-glucoside 4'-O-gentobioside;4,2',4',6'-Tetrahydroxychalcone 2'-O-glucoside 4'-O-gentobioside |
| − | CAS_RN 329272-53-9 | + | CAS_RN 329272-53-9 |
| − | FORMULA C33H42O20 | + | FORMULA C33H42O20 |
| − | EXACTMASS 758.226943784 | + | EXACTMASS 758.226943784 |
| − | AVERAGEMASS 758.67458 | + | AVERAGEMASS 758.67458 |
| − | SMILES C(C(C(O)1)OC(OCC(C2O)OC(Oc(c4)cc(OC(O5)C(O)C(O)C(O)C5CO)c(c4O)C(C=Cc(c3)ccc(c3)O)=O)C(C2O)O)C(C1O)O)O | + | SMILES C(C(C(O)1)OC(OCC(C2O)OC(Oc(c4)cc(OC(O5)C(O)C(O)C(O)C5CO)c(c4O)C(C=Cc(c3)ccc(c3)O)=O)C(C2O)O)C(C1O)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
53 57 0 0 0 0 0 0 0 0999 V2000
0.3373 0.4820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3373 -0.3430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0518 -0.7555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7662 -0.3430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7662 0.4820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0518 0.8945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4807 -0.7555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1952 -0.3430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1952 0.4820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9097 0.8945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6241 0.4820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.3386 0.8945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.3386 1.7195 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6241 2.1320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9097 1.7195 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4807 -1.5805 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6331 -1.4358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1552 -2.1086 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3800 -1.8254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4312 -1.9769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0466 -1.3039 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8218 -1.5870 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0331 -1.0185 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0688 -1.9622 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6262 -1.8928 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4907 -2.4787 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0518 -1.4667 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3494 0.8785 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6163 -0.7038 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3375 0.8118 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.0198 2.1128 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8306 1.0078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4180 0.2931 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6197 0.5023 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8262 0.2754 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2387 0.9902 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0371 0.7811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1941 1.3669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6740 1.6440 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3138 0.5246 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8667 0.5522 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7911 -0.1377 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.5366 1.8425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.1240 1.1278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3257 1.3370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5322 1.1101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9447 1.8249 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.7431 1.6158 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9001 2.2016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.3800 2.4787 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-6.0198 1.3593 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.5727 1.3869 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.4971 0.6970 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 11 2 0 0 0 0
11 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 10 1 0 0 0 0
7 16 2 0 0 0 0
17 18 1 1 0 0 0
18 19 1 1 0 0 0
20 19 1 1 0 0 0
20 21 1 0 0 0 0
21 22 1 0 0 0 0
22 17 1 0 0 0 0
22 23 1 0 0 0 0
17 24 1 0 0 0 0
18 25 1 0 0 0 0
19 26 1 0 0 0 0
3 27 1 0 0 0 0
1 28 1 0 0 0 0
29 23 1 0 0 0 0
5 30 1 0 0 0 0
27 20 1 0 0 0 0
13 31 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
37 38 1 0 0 0 0
38 39 1 0 0 0 0
32 40 1 0 0 0 0
33 41 1 0 0 0 0
34 42 1 0 0 0 0
43 44 1 1 0 0 0
44 45 1 1 0 0 0
46 45 1 1 0 0 0
46 47 1 0 0 0 0
47 48 1 0 0 0 0
48 43 1 0 0 0 0
48 49 1 0 0 0 0
49 50 1 0 0 0 0
43 51 1 0 0 0 0
44 52 1 0 0 0 0
45 53 1 0 0 0 0
46 39 1 0 0 0 0
28 35 1 0 0 0 0
S SKP 8
ID FL1CAAGS0009
KNApSAcK_ID C00014505
NAME Chalconaringenin 2'-O-glucoside 4'-O-gentobioside;4,2',4',6'-Tetrahydroxychalcone 2'-O-glucoside 4'-O-gentobioside
CAS_RN 329272-53-9
FORMULA C33H42O20
EXACTMASS 758.226943784
AVERAGEMASS 758.67458
SMILES C(C(C(O)1)OC(OCC(C2O)OC(Oc(c4)cc(OC(O5)C(O)C(O)C(O)C5CO)c(c4O)C(C=Cc(c3)ccc(c3)O)=O)C(C2O)O)C(C1O)O)O
M END