Mol:FL1CAAGS0002
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 45 0 0 0 0 0 0 0 0999 V2000 | + | 42 45 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.1093 1.1192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1093 1.1192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1093 0.4795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1093 0.4795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6633 0.1596 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6633 0.1596 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2173 0.4795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2173 0.4795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2173 1.1192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2173 1.1192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6633 1.4390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6633 1.4390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7710 0.1598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7710 0.1598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3236 0.4788 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3236 0.4788 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8749 0.1605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8749 0.1605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4250 0.4781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4250 0.4781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9634 0.1673 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9634 0.1673 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5018 0.4781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5018 0.4781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5018 1.0998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5018 1.0998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9634 1.4106 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9634 1.4106 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4250 1.0998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4250 1.0998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7710 -0.4782 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7710 -0.4782 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0270 1.4030 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0270 1.4030 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6633 -0.4797 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6633 -0.4797 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4444 1.4389 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4444 1.4389 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7710 1.4389 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7710 1.4389 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2326 -0.6963 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2326 -0.6963 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8864 -1.1534 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8864 -1.1534 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3878 -0.9595 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3878 -0.9595 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1319 -0.9652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1319 -0.9652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2563 -0.6046 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2563 -0.6046 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7656 -0.7875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7656 -0.7875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6197 -0.9198 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6197 -0.9198 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4261 -1.1796 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4261 -1.1796 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1021 -1.4390 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1021 -1.4390 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0010 -0.4689 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0010 -0.4689 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0010 0.2529 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0010 0.2529 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6188 0.6096 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6188 0.6096 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6188 1.3219 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6188 1.3219 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2356 0.2535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2356 0.2535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8525 0.6096 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8525 0.6096 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4693 0.2535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4693 0.2535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9885 0.5533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9885 0.5533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5078 0.2535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5078 0.2535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5078 -0.3461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5078 -0.3461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9885 -0.6459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9885 -0.6459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4693 -0.3461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4693 -0.3461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0270 -0.6458 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0270 -0.6458 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 11 2 0 0 0 0 | + | 10 11 2 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 10 1 0 0 0 0 | + | 15 10 1 0 0 0 0 |
| − | 7 16 2 0 0 0 0 | + | 7 16 2 0 0 0 0 |
| − | 17 13 1 0 0 0 0 | + | 17 13 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 5 20 1 0 0 0 0 | + | 5 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 24 18 1 0 0 0 0 | + | 24 18 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 2 0 0 0 0 | + | 32 33 2 0 0 0 0 |
| − | 32 34 1 0 0 0 0 | + | 32 34 1 0 0 0 0 |
| − | 34 35 2 0 0 0 0 | + | 34 35 2 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 2 0 0 0 0 | + | 40 41 2 0 0 0 0 |
| − | 41 36 1 0 0 0 0 | + | 41 36 1 0 0 0 0 |
| − | 39 42 1 0 0 0 0 | + | 39 42 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL1CAAGS0002 | + | ID FL1CAAGS0002 |
| − | KNApSAcK_ID C00007879 | + | KNApSAcK_ID C00007879 |
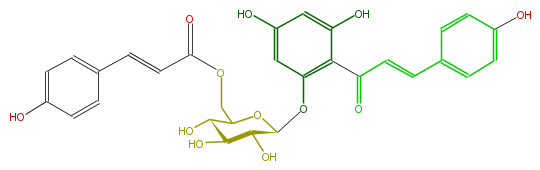
| − | NAME Chalconaringenin 2'-(6''-p-coumarylglucoside) | + | NAME Chalconaringenin 2'-(6''-p-coumarylglucoside) |
| − | CAS_RN 27960-53-8 | + | CAS_RN 27960-53-8 |
| − | FORMULA C30H28O12 | + | FORMULA C30H28O12 |
| − | EXACTMASS 580.15807636 | + | EXACTMASS 580.15807636 |
| − | AVERAGEMASS 580.53612 | + | AVERAGEMASS 580.53612 |
| − | SMILES OC(C3O)C(COC(C=Cc(c4)ccc(O)c4)=O)OC(C3O)Oc(c1C(=O)C=Cc(c2)ccc(O)c2)cc(O)cc1O | + | SMILES OC(C3O)C(COC(C=Cc(c4)ccc(O)c4)=O)OC(C3O)Oc(c1C(=O)C=Cc(c2)ccc(O)c2)cc(O)cc1O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 45 0 0 0 0 0 0 0 0999 V2000
0.1093 1.1192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1093 0.4795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6633 0.1596 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2173 0.4795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2173 1.1192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6633 1.4390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7710 0.1598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3236 0.4788 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8749 0.1605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4250 0.4781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9634 0.1673 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5018 0.4781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5018 1.0998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9634 1.4106 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4250 1.0998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7710 -0.4782 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.0270 1.4030 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6633 -0.4797 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4444 1.4389 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7710 1.4389 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2326 -0.6963 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8864 -1.1534 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3878 -0.9595 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1319 -0.9652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2563 -0.6046 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7656 -0.7875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6197 -0.9198 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4261 -1.1796 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1021 -1.4390 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0010 -0.4689 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0010 0.2529 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6188 0.6096 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6188 1.3219 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2356 0.2535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8525 0.6096 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4693 0.2535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9885 0.5533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5078 0.2535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5078 -0.3461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9885 -0.6459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4693 -0.3461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0270 -0.6458 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 11 2 0 0 0 0
11 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 10 1 0 0 0 0
7 16 2 0 0 0 0
17 13 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
5 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
26 30 1 0 0 0 0
24 18 1 0 0 0 0
30 31 1 0 0 0 0
31 32 1 0 0 0 0
32 33 2 0 0 0 0
32 34 1 0 0 0 0
34 35 2 0 0 0 0
35 36 1 0 0 0 0
36 37 2 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
39 40 1 0 0 0 0
40 41 2 0 0 0 0
41 36 1 0 0 0 0
39 42 1 0 0 0 0
S SKP 8
ID FL1CAAGS0002
KNApSAcK_ID C00007879
NAME Chalconaringenin 2'-(6''-p-coumarylglucoside)
CAS_RN 27960-53-8
FORMULA C30H28O12
EXACTMASS 580.15807636
AVERAGEMASS 580.53612
SMILES OC(C3O)C(COC(C=Cc(c4)ccc(O)c4)=O)OC(C3O)Oc(c1C(=O)C=Cc(c2)ccc(O)c2)cc(O)cc1O
M END