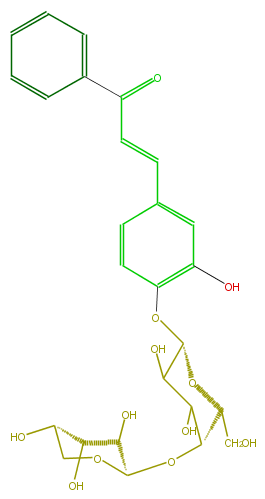
Mol:FL1C9CGS0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 38 41 0 0 0 0 0 0 0 0999 V2000 | + | 38 41 0 0 0 0 0 0 0 0999 V2000 |
| − | -5.0902 -0.8575 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0902 -0.8575 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0902 -1.7064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0902 -1.7064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3549 -2.1311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3549 -2.1311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6195 -1.7064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6195 -1.7064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6195 -0.8575 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6195 -0.8575 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3549 -0.4331 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3549 -0.4331 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8846 -2.1308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8846 -2.1308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1513 -1.7074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1513 -1.7074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4195 -2.1299 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4195 -2.1299 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6893 -1.7084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6893 -1.7084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0252 -2.1210 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0252 -2.1210 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7398 -1.7084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7398 -1.7084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7398 -0.8833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7398 -0.8833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0252 -0.4707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0252 -0.4707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6893 -0.8833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6893 -0.8833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8846 -2.9776 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8846 -2.9776 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3881 -0.0510 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3881 -0.0510 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9888 -0.7425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9888 -0.7425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7566 -0.5231 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7566 -0.5231 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5291 -0.7425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5291 -0.7425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9285 -0.0510 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9285 -0.0510 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1605 -0.2704 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1605 -0.2704 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7941 -0.2102 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7941 -0.2102 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5039 0.0730 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5039 0.0730 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9111 0.6322 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9111 0.6322 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2630 -0.5481 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2630 -0.5481 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0527 1.3636 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0527 1.3636 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7168 1.5415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7168 1.5415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1158 1.8754 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1158 1.8754 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7791 2.4793 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7791 2.4793 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1150 2.3014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1150 2.3014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7160 1.9674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7160 1.9674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9760 2.9776 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9760 2.9776 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3324 2.5837 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3324 2.5837 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6408 0.9518 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6408 0.9518 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4542 -2.1209 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4542 -2.1209 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1418 -0.5421 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1418 -0.5421 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0902 -1.0896 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0902 -1.0896 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 11 2 0 0 0 0 | + | 10 11 2 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 10 1 0 0 0 0 | + | 15 10 1 0 0 0 0 |
| − | 7 16 2 0 0 0 0 | + | 7 16 2 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 19 1 1 0 0 0 | + | 18 19 1 1 0 0 0 |
| − | 19 20 1 1 0 0 0 | + | 19 20 1 1 0 0 0 |
| − | 21 20 1 1 0 0 0 | + | 21 20 1 1 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 17 1 0 0 0 0 | + | 22 17 1 0 0 0 0 |
| − | 17 23 1 0 0 0 0 | + | 17 23 1 0 0 0 0 |
| − | 22 24 1 0 0 0 0 | + | 22 24 1 0 0 0 0 |
| − | 21 25 1 0 0 0 0 | + | 21 25 1 0 0 0 0 |
| − | 18 26 1 0 0 0 0 | + | 18 26 1 0 0 0 0 |
| − | 26 13 1 0 0 0 0 | + | 26 13 1 0 0 0 0 |
| − | 28 27 1 1 0 0 0 | + | 28 27 1 1 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 27 1 1 0 0 0 | + | 32 27 1 1 0 0 0 |
| − | 31 33 1 0 0 0 0 | + | 31 33 1 0 0 0 0 |
| − | 32 34 1 0 0 0 0 | + | 32 34 1 0 0 0 0 |
| − | 27 35 1 0 0 0 0 | + | 27 35 1 0 0 0 0 |
| − | 25 28 1 0 0 0 0 | + | 25 28 1 0 0 0 0 |
| − | 12 36 1 0 0 0 0 | + | 12 36 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 20 37 1 0 0 0 0 | + | 20 37 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 37 38 | + | M SAL 1 2 37 38 |
| − | M SBL 1 1 41 | + | M SBL 1 1 41 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 41 -0.6127 -0.2004 | + | M SBV 1 41 -0.6127 -0.2004 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL1C9CGS0001 | + | ID FL1C9CGS0001 |
| − | FORMULA C26H30O12 | + | FORMULA C26H30O12 |
| − | EXACTMASS 534.173726424 | + | EXACTMASS 534.173726424 |
| − | AVERAGEMASS 534.5092 | + | AVERAGEMASS 534.5092 |
| − | SMILES c(c1)ccc(C(C=Cc(c4)cc(O)c(c4)OC(C3O)OC(C(C(O)3)OC(C(O)2)OCC(O)C2O)CO)=O)c1 | + | SMILES c(c1)ccc(C(C=Cc(c4)cc(O)c(c4)OC(C3O)OC(C(C(O)3)OC(C(O)2)OCC(O)C2O)CO)=O)c1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
38 41 0 0 0 0 0 0 0 0999 V2000
-5.0902 -0.8575 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0902 -1.7064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3549 -2.1311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6195 -1.7064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6195 -0.8575 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3549 -0.4331 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8846 -2.1308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1513 -1.7074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4195 -2.1299 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6893 -1.7084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0252 -2.1210 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7398 -1.7084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7398 -0.8833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0252 -0.4707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6893 -0.8833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8846 -2.9776 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3881 -0.0510 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9888 -0.7425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7566 -0.5231 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5291 -0.7425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9285 -0.0510 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1605 -0.2704 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7941 -0.2102 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5039 0.0730 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9111 0.6322 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2630 -0.5481 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0527 1.3636 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7168 1.5415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1158 1.8754 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7791 2.4793 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1150 2.3014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7160 1.9674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9760 2.9776 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3324 2.5837 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6408 0.9518 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4542 -2.1209 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1418 -0.5421 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.0902 -1.0896 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 11 2 0 0 0 0
11 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 10 1 0 0 0 0
7 16 2 0 0 0 0
17 18 1 0 0 0 0
18 19 1 1 0 0 0
19 20 1 1 0 0 0
21 20 1 1 0 0 0
21 22 1 0 0 0 0
22 17 1 0 0 0 0
17 23 1 0 0 0 0
22 24 1 0 0 0 0
21 25 1 0 0 0 0
18 26 1 0 0 0 0
26 13 1 0 0 0 0
28 27 1 1 0 0 0
28 29 1 0 0 0 0
29 30 1 0 0 0 0
30 31 1 0 0 0 0
31 32 1 1 0 0 0
32 27 1 1 0 0 0
31 33 1 0 0 0 0
32 34 1 0 0 0 0
27 35 1 0 0 0 0
25 28 1 0 0 0 0
12 36 1 0 0 0 0
37 38 1 0 0 0 0
20 37 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 37 38
M SBL 1 1 41
M SMT 1 CH2OH
M SBV 1 41 -0.6127 -0.2004
S SKP 5
ID FL1C9CGS0001
FORMULA C26H30O12
EXACTMASS 534.173726424
AVERAGEMASS 534.5092
SMILES c(c1)ccc(C(C=Cc(c4)cc(O)c(c4)OC(C3O)OC(C(C(O)3)OC(C(O)2)OCC(O)C2O)CO)=O)c1
M END