Mol:FL1C1CNI0002
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 40 41 0 0 0 0 0 0 0 0999 V2000 | + | 40 41 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.0108 -0.3678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0108 -0.3678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0108 -1.1928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0108 -1.1928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7037 -1.6053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7037 -1.6053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4181 -1.1928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4181 -1.1928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4181 -0.3678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4181 -0.3678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7037 0.0447 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7037 0.0447 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1326 -1.6053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1326 -1.6053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1326 -2.4303 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1326 -2.4303 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8471 -1.1928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8471 -1.1928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8471 -0.3678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8471 -0.3678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5616 0.0447 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5616 0.0447 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2759 -0.3894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2759 -0.3894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9933 0.0068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9933 0.0068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0121 0.8317 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0121 0.8317 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3009 1.2604 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3009 1.2604 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5802 0.8588 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5802 0.8588 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.7266 1.2442 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.7266 1.2442 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7037 -2.4303 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7037 -2.4303 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7253 0.0447 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7253 0.0447 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.7078 -0.4057 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.7078 -0.4057 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7253 -1.6053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7253 -1.6053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4397 -1.1928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4397 -1.1928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1542 -1.6053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1542 -1.6053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1542 -2.4303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1542 -2.4303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8687 -1.1928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8687 -1.1928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5831 -1.6053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5831 -1.6053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2976 -1.1928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2976 -1.1928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0121 -1.6053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0121 -1.6053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0121 -2.4303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0121 -2.4303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.7266 -1.1928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.7266 -1.1928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2907 2.0178 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2907 2.0178 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5763 2.4303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5763 2.4303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8618 2.0178 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8618 2.0178 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8618 1.1928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8618 1.1928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1473 2.4303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1473 2.4303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4329 2.0178 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4329 2.0178 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7184 2.4303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7184 2.4303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0039 2.0178 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0039 2.0178 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0039 1.1928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0039 1.1928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7106 2.4303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7106 2.4303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 7 9 1 0 0 0 0 | + | 7 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 11 1 0 0 0 0 | + | 10 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 14 17 1 0 0 0 0 | + | 14 17 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 13 20 1 0 0 0 0 | + | 13 20 1 0 0 0 0 |
| − | 2 21 1 0 0 0 0 | + | 2 21 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 2 0 0 0 0 | + | 22 23 2 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 23 25 1 0 0 0 0 | + | 23 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 2 0 0 0 0 | + | 27 28 2 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 28 30 1 0 0 0 0 | + | 28 30 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 2 0 0 0 0 | + | 32 33 2 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 33 35 1 0 0 0 0 | + | 33 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 2 0 0 0 0 | + | 37 38 2 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 38 40 1 0 0 0 0 | + | 38 40 1 0 0 0 0 |
| − | 15 31 1 0 0 0 0 | + | 15 31 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL1C1CNI0002 | + | ID FL1C1CNI0002 |
| − | KNApSAcK_ID C00014485 | + | KNApSAcK_ID C00014485 |
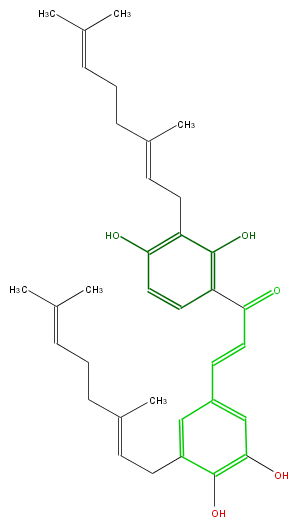
| − | NAME 5,3'-Digeranyl-3,4,2',4'-tetrahydroxychalcone;Prorepensin | + | NAME 5,3'-Digeranyl-3,4,2',4'-tetrahydroxychalcone;Prorepensin |
| − | CAS_RN 441772-63-0 | + | CAS_RN 441772-63-0 |
| − | FORMULA C35H44O5 | + | FORMULA C35H44O5 |
| − | EXACTMASS 544.318874518 | + | EXACTMASS 544.318874518 |
| − | AVERAGEMASS 544.72086 | + | AVERAGEMASS 544.72086 |
| − | SMILES c(CC=C(C)CCC=C(C)C)(c1)c(c(O)cc1C=CC(c(c2O)ccc(O)c2CC=C(CCC=C(C)C)C)=O)O | + | SMILES c(CC=C(C)CCC=C(C)C)(c1)c(c(O)cc1C=CC(c(c2O)ccc(O)c2CC=C(CCC=C(C)C)C)=O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
40 41 0 0 0 0 0 0 0 0999 V2000
-0.0108 -0.3678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0108 -1.1928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7037 -1.6053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4181 -1.1928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4181 -0.3678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7037 0.0447 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1326 -1.6053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1326 -2.4303 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8471 -1.1928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8471 -0.3678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5616 0.0447 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2759 -0.3894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9933 0.0068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.0121 0.8317 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3009 1.2604 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5802 0.8588 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.7266 1.2442 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7037 -2.4303 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7253 0.0447 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.7078 -0.4057 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7253 -1.6053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4397 -1.1928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1542 -1.6053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1542 -2.4303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8687 -1.1928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5831 -1.6053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2976 -1.1928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0121 -1.6053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0121 -2.4303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.7266 -1.1928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2907 2.0178 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5763 2.4303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8618 2.0178 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8618 1.1928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1473 2.4303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4329 2.0178 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7184 2.4303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0039 2.0178 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0039 1.1928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7106 2.4303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
7 9 1 0 0 0 0
9 10 2 0 0 0 0
10 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
14 17 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
13 20 1 0 0 0 0
2 21 1 0 0 0 0
21 22 1 0 0 0 0
22 23 2 0 0 0 0
23 24 1 0 0 0 0
23 25 1 0 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 28 2 0 0 0 0
28 29 1 0 0 0 0
28 30 1 0 0 0 0
31 32 1 0 0 0 0
32 33 2 0 0 0 0
33 34 1 0 0 0 0
33 35 1 0 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 38 2 0 0 0 0
38 39 1 0 0 0 0
38 40 1 0 0 0 0
15 31 1 0 0 0 0
S SKP 8
ID FL1C1CNI0002
KNApSAcK_ID C00014485
NAME 5,3'-Digeranyl-3,4,2',4'-tetrahydroxychalcone;Prorepensin
CAS_RN 441772-63-0
FORMULA C35H44O5
EXACTMASS 544.318874518
AVERAGEMASS 544.72086
SMILES c(CC=C(C)CCC=C(C)C)(c1)c(c(O)cc1C=CC(c(c2O)ccc(O)c2CC=C(CCC=C(C)C)C)=O)O
M END