Mol:FL1C1AGS0005
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 41 44 0 0 0 0 0 0 0 0999 V2000 | + | 41 44 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.0983 -0.6365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0983 -0.6365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0983 -1.2762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0983 -1.2762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5443 -1.5960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5443 -1.5960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0097 -1.2762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0097 -1.2762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0097 -0.6365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0097 -0.6365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5443 -0.3166 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5443 -0.3166 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5635 -1.5959 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5635 -1.5959 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1160 -1.2769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1160 -1.2769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6673 -1.5952 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6673 -1.5952 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2175 -1.2776 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2175 -1.2776 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7558 -1.5884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7558 -1.5884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2942 -1.2776 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2942 -1.2776 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2942 -0.6559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2942 -0.6559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7558 -0.3451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7558 -0.3451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2175 -0.6559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2175 -0.6559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5635 -2.2339 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5635 -2.2339 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8194 -0.3527 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8194 -0.3527 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5443 -2.2354 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5443 -2.2354 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6520 -0.3168 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6520 -0.3168 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4451 0.6082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4451 0.6082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0130 0.2803 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 4.0130 0.2803 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.8328 0.9108 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 3.8328 0.9108 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 4.0130 1.5452 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 4.0130 1.5452 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 3.4451 1.8731 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.4451 1.8731 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.6253 1.2425 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 3.6253 1.2425 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 3.4004 2.3835 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4004 2.3835 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3674 1.7498 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3674 1.7498 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2504 0.6697 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2504 0.6697 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8633 -0.5339 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -3.8633 -0.5339 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -3.5171 -0.9909 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.5171 -0.9909 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -3.0185 -0.7970 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.0185 -0.7970 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.4988 -0.8027 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.4988 -0.8027 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.8870 -0.4421 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8870 -0.4421 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3963 -0.6250 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -3.3963 -0.6250 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -4.2503 -0.3104 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2503 -0.3104 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8439 -1.4211 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8439 -1.4211 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7328 -1.2766 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7328 -1.2766 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9491 1.4608 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9491 1.4608 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2072 0.7902 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2072 0.7902 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6659 0.1020 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6659 0.1020 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0278 0.8719 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0278 0.8719 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 11 2 0 0 0 0 | + | 10 11 2 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 10 1 0 0 0 0 | + | 15 10 1 0 0 0 0 |
| − | 7 16 2 0 0 0 0 | + | 7 16 2 0 0 0 0 |
| − | 17 13 1 0 0 0 0 | + | 17 13 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 21 20 1 1 0 0 0 | + | 21 20 1 1 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 25 20 1 1 0 0 0 | + | 25 20 1 1 0 0 0 |
| − | 24 26 1 0 0 0 0 | + | 24 26 1 0 0 0 0 |
| − | 23 27 1 0 0 0 0 | + | 23 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 21 17 1 0 0 0 0 | + | 21 17 1 0 0 0 0 |
| − | 29 30 1 1 0 0 0 | + | 29 30 1 1 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 29 1 0 0 0 0 | + | 34 29 1 0 0 0 0 |
| − | 29 35 1 0 0 0 0 | + | 29 35 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 19 1 0 0 0 0 | + | 32 19 1 0 0 0 0 |
| − | 25 38 1 0 0 0 0 | + | 25 38 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 40 41 | + | M SAL 2 2 40 41 |
| − | M SBL 2 1 43 | + | M SBL 2 1 43 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 43 -3.6659 0.102 | + | M SVB 2 43 -3.6659 0.102 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 38 39 | + | M SAL 1 2 38 39 |
| − | M SBL 1 1 41 | + | M SBL 1 1 41 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SVB 1 41 3.1047 1.7261 | + | M SVB 1 41 3.1047 1.7261 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL1C1AGS0005 | + | ID FL1C1AGS0005 |
| − | KNApSAcK_ID C00007187 | + | KNApSAcK_ID C00007187 |
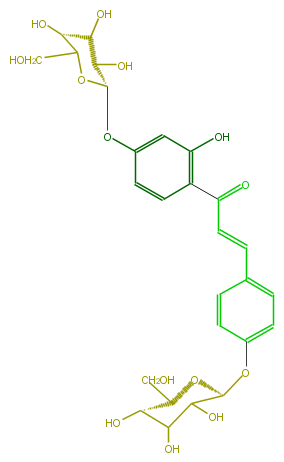
| − | NAME Isoliquiritigenin 4,4'-diglucoside | + | NAME Isoliquiritigenin 4,4'-diglucoside |
| − | CAS_RN 69262-36-8 | + | CAS_RN 69262-36-8 |
| − | FORMULA C27H32O14 | + | FORMULA C27H32O14 |
| − | EXACTMASS 580.179205732 | + | EXACTMASS 580.179205732 |
| − | AVERAGEMASS 580.53458 | + | AVERAGEMASS 580.53458 |
| − | SMILES [C@H]([C@@H](O)4)(CO)O[C@@H](C(O)C(O)4)Oc(c1)ccc(C=CC(c(c(O)2)ccc(O[C@@H]([C@H]3O)OC([C@H](O)[C@@H]3O)CO)c2)=O)c1 | + | SMILES [C@H]([C@@H](O)4)(CO)O[C@@H](C(O)C(O)4)Oc(c1)ccc(C=CC(c(c(O)2)ccc(O[C@@H]([C@H]3O)OC([C@H](O)[C@@H]3O)CO)c2)=O)c1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
41 44 0 0 0 0 0 0 0 0999 V2000
-1.0983 -0.6365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0983 -1.2762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5443 -1.5960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0097 -1.2762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0097 -0.6365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5443 -0.3166 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5635 -1.5959 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1160 -1.2769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6673 -1.5952 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2175 -1.2776 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7558 -1.5884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2942 -1.2776 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2942 -0.6559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7558 -0.3451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2175 -0.6559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5635 -2.2339 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8194 -0.3527 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5443 -2.2354 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6520 -0.3168 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4451 0.6082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0130 0.2803 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.8328 0.9108 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
4.0130 1.5452 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
3.4451 1.8731 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.6253 1.2425 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
3.4004 2.3835 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.3674 1.7498 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2504 0.6697 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8633 -0.5339 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-3.5171 -0.9909 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-3.0185 -0.7970 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.4988 -0.8027 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.8870 -0.4421 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3963 -0.6250 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-4.2503 -0.3104 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8439 -1.4211 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7328 -1.2766 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9491 1.4608 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2072 0.7902 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6659 0.1020 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0278 0.8719 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 11 2 0 0 0 0
11 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 10 1 0 0 0 0
7 16 2 0 0 0 0
17 13 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
21 20 1 1 0 0 0
21 22 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 25 1 1 0 0 0
25 20 1 1 0 0 0
24 26 1 0 0 0 0
23 27 1 0 0 0 0
22 28 1 0 0 0 0
21 17 1 0 0 0 0
29 30 1 1 0 0 0
30 31 1 1 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 29 1 0 0 0 0
29 35 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 19 1 0 0 0 0
25 38 1 0 0 0 0
38 39 1 0 0 0 0
34 40 1 0 0 0 0
40 41 1 0 0 0 0
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 40 41
M SBL 2 1 43
M SMT 2 CH2OH
M SVB 2 43 -3.6659 0.102
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 38 39
M SBL 1 1 41
M SMT 1 CH2OH
M SVB 1 41 3.1047 1.7261
S SKP 8
ID FL1C1AGS0005
KNApSAcK_ID C00007187
NAME Isoliquiritigenin 4,4'-diglucoside
CAS_RN 69262-36-8
FORMULA C27H32O14
EXACTMASS 580.179205732
AVERAGEMASS 580.53458
SMILES [C@H]([C@@H](O)4)(CO)O[C@@H](C(O)C(O)4)Oc(c1)ccc(C=CC(c(c(O)2)ccc(O[C@@H]([C@H]3O)OC([C@H](O)[C@@H]3O)CO)c2)=O)c1
M END