Mol:FLNACCGS0004
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.8879 1.3542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8879 1.3542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6073 1.7695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6073 1.7695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3265 1.3544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3265 1.3544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3265 0.4931 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3265 0.4931 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0723 0.0625 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0723 0.0625 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8182 0.4931 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8182 0.4931 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8182 1.3544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8182 1.3544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0723 1.7849 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0723 1.7849 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8879 0.5239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8879 0.5239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6073 0.1088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6073 0.1088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0723 -0.7981 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0723 -0.7981 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3572 -1.2109 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3572 -1.2109 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3572 -2.0366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3572 -2.0366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0723 -2.4494 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0723 -2.4494 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7874 -2.0366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7874 -2.0366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7874 -1.2109 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7874 -1.2109 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5634 1.7846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5634 1.7846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0723 -3.2744 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0723 -3.2744 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6102 -0.7337 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6102 -0.7337 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8348 -0.7109 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8348 -0.7109 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3715 -1.3224 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3715 -1.3224 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2955 -1.0630 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2955 -1.0630 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9392 -1.0560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9392 -1.0560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4715 -0.5882 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4715 -0.5882 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2098 -0.8330 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2098 -0.8330 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3489 -0.8886 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3489 -0.8886 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9878 -1.2985 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9878 -1.2985 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9088 -1.5024 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9088 -1.5024 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7716 -0.2572 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7716 -0.2572 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5619 -0.2583 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5619 -0.2583 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5018 -2.4491 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5018 -2.4491 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9910 -0.0898 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9910 -0.0898 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5829 -0.7393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5829 -0.7393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8957 -0.5390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8957 -0.5390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2539 -0.5881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2539 -0.5881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6790 -0.0814 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6790 -0.0814 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3792 -0.2657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3792 -0.2657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5634 -0.2214 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5634 -0.2214 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2007 -0.6921 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2007 -0.6921 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2902 -0.9281 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2902 -0.9281 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4259 2.1544 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4259 2.1544 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2205 3.2744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2205 3.2744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 7 1 0 0 0 0 | + | 6 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 3 1 0 0 0 0 | + | 8 3 1 0 0 0 0 |
| − | 1 9 1 0 0 0 0 | + | 1 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 4 1 0 0 0 0 | + | 10 4 1 0 0 0 0 |
| − | 5 11 1 0 0 0 0 | + | 5 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 7 17 2 0 0 0 0 | + | 7 17 2 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 10 19 1 0 0 0 0 | + | 10 19 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 23 19 1 0 0 0 0 | + | 23 19 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 15 31 1 0 0 0 0 | + | 15 31 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 1 41 1 0 0 0 0 | + | 1 41 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 41 42 | + | M SAL 1 2 41 42 |
| − | M SBL 1 1 46 | + | M SBL 1 1 46 |
| − | M SMT 1 ^ OCH3 | + | M SMT 1 ^ OCH3 |
| − | M SBV 1 46 0.4619 -0.8002 | + | M SBV 1 46 0.4619 -0.8002 |
| − | S SKP 5 | + | S SKP 5 |
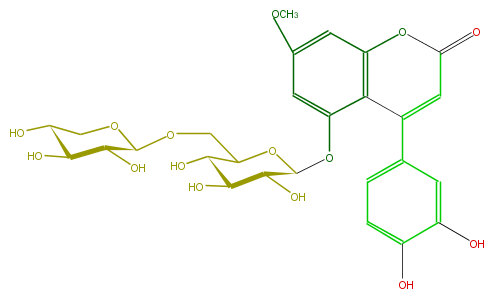
| − | ID FLNACCGS0004 | + | ID FLNACCGS0004 |
| − | FORMULA C27H30O15 | + | FORMULA C27H30O15 |
| − | EXACTMASS 594.15847029 | + | EXACTMASS 594.15847029 |
| − | AVERAGEMASS 594.5181 | + | AVERAGEMASS 594.5181 |
| − | SMILES C(Oc(c43)cc(OC)cc3OC(C=C(c(c5)cc(O)c(O)c5)4)=O)(C(O)2)OC(C(O)C2O)COC(O1)C(O)C(C(O)C1)O | + | SMILES C(Oc(c43)cc(OC)cc3OC(C=C(c(c5)cc(O)c(O)c5)4)=O)(C(O)2)OC(C(O)C2O)COC(O1)C(O)C(C(O)C1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
0.8879 1.3542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6073 1.7695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3265 1.3544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3265 0.4931 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0723 0.0625 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8182 0.4931 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8182 1.3544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0723 1.7849 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8879 0.5239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6073 0.1088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0723 -0.7981 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3572 -1.2109 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3572 -2.0366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0723 -2.4494 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7874 -2.0366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7874 -1.2109 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5634 1.7846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0723 -3.2744 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6102 -0.7337 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8348 -0.7109 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3715 -1.3224 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2955 -1.0630 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9392 -1.0560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4715 -0.5882 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2098 -0.8330 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3489 -0.8886 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9878 -1.2985 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9088 -1.5024 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7716 -0.2572 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5619 -0.2583 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5018 -2.4491 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9910 -0.0898 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5829 -0.7393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8957 -0.5390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2539 -0.5881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6790 -0.0814 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3792 -0.2657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5634 -0.2214 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2007 -0.6921 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2902 -0.9281 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4259 2.1544 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2205 3.2744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 7 1 0 0 0 0
7 8 1 0 0 0 0
8 3 1 0 0 0 0
1 9 1 0 0 0 0
9 10 2 0 0 0 0
10 4 1 0 0 0 0
5 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
7 17 2 0 0 0 0
14 18 1 0 0 0 0
10 19 1 0 0 0 0
20 21 1 1 0 0 0
21 22 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 20 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
25 29 1 0 0 0 0
23 19 1 0 0 0 0
29 30 1 0 0 0 0
15 31 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 40 1 0 0 0 0
35 30 1 0 0 0 0
41 42 1 0 0 0 0
1 41 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 41 42
M SBL 1 1 46
M SMT 1 ^ OCH3
M SBV 1 46 0.4619 -0.8002
S SKP 5
ID FLNACCGS0004
FORMULA C27H30O15
EXACTMASS 594.15847029
AVERAGEMASS 594.5181
SMILES C(Oc(c43)cc(OC)cc3OC(C=C(c(c5)cc(O)c(O)c5)4)=O)(C(O)2)OC(C(O)C2O)COC(O1)C(O)C(C(O)C1)O
M END