Mol:FL7AACGL0059
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 61 66 0 0 0 0 0 0 0 0999 V2000 | + | 61 66 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.1306 -0.4336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1306 -0.4336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1306 -1.0759 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1306 -1.0759 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5743 -1.3971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5743 -1.3971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0180 -1.0759 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0180 -1.0759 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0180 -0.4336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0180 -0.4336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5743 -0.1124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5743 -0.1124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4617 -1.3971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4617 -1.3971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9054 -1.0759 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9054 -1.0759 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9054 -0.4336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9054 -0.4336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4617 -0.1124 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 | + | -1.4617 -0.1124 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3493 -0.1125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3493 -0.1125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2177 -0.4399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2177 -0.4399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7847 -0.1125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7847 -0.1125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7847 0.5422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7847 0.5422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2177 0.8695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2177 0.8695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3493 0.5422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3493 0.5422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6867 -0.1125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6867 -0.1125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3515 0.8694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3515 0.8694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5743 -2.0393 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5743 -2.0393 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4893 -1.7967 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4893 -1.7967 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2177 1.5240 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2177 1.5240 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7713 -1.4553 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7713 -1.4553 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4704 -1.9764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4704 -1.9764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0489 -1.8110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0489 -1.8110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6309 -1.9764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6309 -1.9764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9318 -1.4553 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9318 -1.4553 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3533 -1.6206 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3533 -1.6206 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2126 -1.6050 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2126 -1.6050 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5914 -1.2080 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5914 -1.2080 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6351 -1.6438 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6351 -1.6438 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1977 -1.9805 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1977 -1.9805 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5895 -2.9911 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5895 -2.9911 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5792 -3.6923 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5792 -3.6923 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9639 -4.2121 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9639 -4.2121 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0310 2.5848 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0310 2.5848 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5369 2.2569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5369 2.2569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3567 2.8875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3567 2.8875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5369 3.5218 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5369 3.5218 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0310 3.8498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0310 3.8498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1492 3.2192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1492 3.2192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3313 4.2121 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3313 4.2121 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5369 3.9310 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5369 3.9310 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5145 3.6089 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5145 3.6089 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7744 2.6463 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7744 2.6463 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1061 3.0789 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1061 3.0789 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1061 2.3300 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1061 2.3300 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6546 2.0693 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6546 2.0693 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7960 2.3300 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7960 2.3300 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1509 1.7154 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1509 1.7154 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7643 1.7154 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7643 1.7154 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0858 2.2723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0858 2.2723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7289 2.2723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7289 2.2723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0504 1.7154 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0504 1.7154 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7289 1.1585 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7289 1.1585 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0858 1.1585 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0858 1.1585 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6925 1.7154 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6925 1.7154 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0499 2.8284 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0499 2.8284 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3063 -4.0364 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3063 -4.0364 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9667 -3.5796 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9667 -3.5796 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6925 -3.9231 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6925 -3.9231 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9667 -2.9498 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9667 -2.9498 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 1 17 1 0 0 0 0 | + | 1 17 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 20 8 1 0 0 0 0 | + | 20 8 1 0 0 0 0 |
| − | 15 21 1 0 0 0 0 | + | 15 21 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 25 31 1 0 0 0 0 | + | 25 31 1 0 0 0 0 |
| − | 23 20 1 0 0 0 0 | + | 23 20 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 33 34 2 0 0 0 0 | + | 33 34 2 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 36 35 1 1 0 0 0 | + | 36 35 1 1 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 39 40 1 1 0 0 0 | + | 39 40 1 1 0 0 0 |
| − | 40 35 1 1 0 0 0 | + | 40 35 1 1 0 0 0 |
| − | 39 41 1 0 0 0 0 | + | 39 41 1 0 0 0 0 |
| − | 38 42 1 0 0 0 0 | + | 38 42 1 0 0 0 0 |
| − | 40 43 1 0 0 0 0 | + | 40 43 1 0 0 0 0 |
| − | 37 44 1 0 0 0 0 | + | 37 44 1 0 0 0 0 |
| − | 36 21 1 0 0 0 0 | + | 36 21 1 0 0 0 0 |
| − | 43 45 1 0 0 0 0 | + | 43 45 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 46 47 2 0 0 0 0 | + | 46 47 2 0 0 0 0 |
| − | 46 48 1 0 0 0 0 | + | 46 48 1 0 0 0 0 |
| − | 48 49 2 0 0 0 0 | + | 48 49 2 0 0 0 0 |
| − | 49 50 1 0 0 0 0 | + | 49 50 1 0 0 0 0 |
| − | 50 51 2 0 0 0 0 | + | 50 51 2 0 0 0 0 |
| − | 51 52 1 0 0 0 0 | + | 51 52 1 0 0 0 0 |
| − | 52 53 2 0 0 0 0 | + | 52 53 2 0 0 0 0 |
| − | 53 54 1 0 0 0 0 | + | 53 54 1 0 0 0 0 |
| − | 54 55 2 0 0 0 0 | + | 54 55 2 0 0 0 0 |
| − | 55 50 1 0 0 0 0 | + | 55 50 1 0 0 0 0 |
| − | 53 56 1 0 0 0 0 | + | 53 56 1 0 0 0 0 |
| − | 52 57 1 0 0 0 0 | + | 52 57 1 0 0 0 0 |
| − | 33 58 1 0 0 0 0 | + | 33 58 1 0 0 0 0 |
| − | 58 59 1 0 0 0 0 | + | 58 59 1 0 0 0 0 |
| − | 59 60 1 0 0 0 0 | + | 59 60 1 0 0 0 0 |
| − | 59 61 2 0 0 0 0 | + | 59 61 2 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL7AACGL0059 | + | ID FL7AACGL0059 |
| − | KNApSAcK_ID C00006830 | + | KNApSAcK_ID C00006830 |
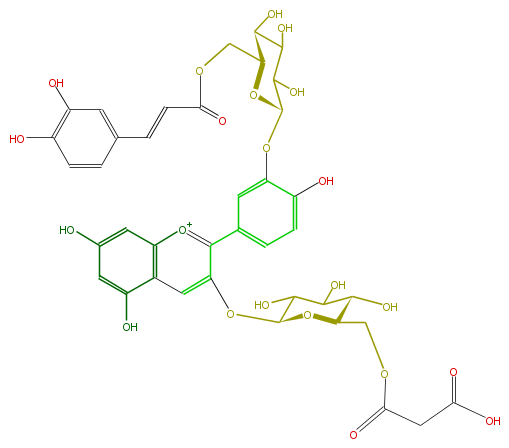
| − | NAME Cyanidin 3-(6''-malonylglucoside-3'-(6'''-caffeylglucoside) | + | NAME Cyanidin 3-(6''-malonylglucoside-3'-(6'''-caffeylglucoside) |
| − | CAS_RN 150447-30-6 | + | CAS_RN 150447-30-6 |
| − | FORMULA C39H39O22 | + | FORMULA C39H39O22 |
| − | EXACTMASS 859.193297932 | + | EXACTMASS 859.193297932 |
| − | AVERAGEMASS 859.7137600000001 | + | AVERAGEMASS 859.7137600000001 |
| − | SMILES c(c13)c(O)cc(c1cc(c(c(c4)cc(OC(C(O)6)OC(C(O)C(O)6)COC(=O)C=Cc(c5)cc(O)c(O)c5)c(O)c4)[o+1]3)OC(C2O)OC(C(C2O)O)COC(=O)CC(O)=O)O | + | SMILES c(c13)c(O)cc(c1cc(c(c(c4)cc(OC(C(O)6)OC(C(O)C(O)6)COC(=O)C=Cc(c5)cc(O)c(O)c5)c(O)c4)[o+1]3)OC(C2O)OC(C(C2O)O)COC(=O)CC(O)=O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
61 66 0 0 0 0 0 0 0 0999 V2000
-3.1306 -0.4336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1306 -1.0759 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5743 -1.3971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0180 -1.0759 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0180 -0.4336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5743 -0.1124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4617 -1.3971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9054 -1.0759 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9054 -0.4336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4617 -0.1124 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
-0.3493 -0.1125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2177 -0.4399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7847 -0.1125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7847 0.5422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2177 0.8695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3493 0.5422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6867 -0.1125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3515 0.8694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5743 -2.0393 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4893 -1.7967 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2177 1.5240 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7713 -1.4553 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4704 -1.9764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0489 -1.8110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6309 -1.9764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9318 -1.4553 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3533 -1.6206 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2126 -1.6050 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5914 -1.2080 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6351 -1.6438 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1977 -1.9805 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5895 -2.9911 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5792 -3.6923 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9639 -4.2121 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0310 2.5848 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5369 2.2569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3567 2.8875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5369 3.5218 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0310 3.8498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1492 3.2192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3313 4.2121 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5369 3.9310 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5145 3.6089 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7744 2.6463 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1061 3.0789 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1061 2.3300 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6546 2.0693 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7960 2.3300 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1509 1.7154 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7643 1.7154 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0858 2.2723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7289 2.2723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0504 1.7154 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7289 1.1585 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0858 1.1585 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6925 1.7154 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0499 2.8284 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3063 -4.0364 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9667 -3.5796 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6925 -3.9231 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9667 -2.9498 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
1 17 1 0 0 0 0
14 18 1 0 0 0 0
3 19 1 0 0 0 0
20 8 1 0 0 0 0
15 21 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
25 31 1 0 0 0 0
23 20 1 0 0 0 0
31 32 1 0 0 0 0
33 34 2 0 0 0 0
32 33 1 0 0 0 0
36 35 1 1 0 0 0
36 37 1 0 0 0 0
37 38 1 0 0 0 0
38 39 1 0 0 0 0
39 40 1 1 0 0 0
40 35 1 1 0 0 0
39 41 1 0 0 0 0
38 42 1 0 0 0 0
40 43 1 0 0 0 0
37 44 1 0 0 0 0
36 21 1 0 0 0 0
43 45 1 0 0 0 0
45 46 1 0 0 0 0
46 47 2 0 0 0 0
46 48 1 0 0 0 0
48 49 2 0 0 0 0
49 50 1 0 0 0 0
50 51 2 0 0 0 0
51 52 1 0 0 0 0
52 53 2 0 0 0 0
53 54 1 0 0 0 0
54 55 2 0 0 0 0
55 50 1 0 0 0 0
53 56 1 0 0 0 0
52 57 1 0 0 0 0
33 58 1 0 0 0 0
58 59 1 0 0 0 0
59 60 1 0 0 0 0
59 61 2 0 0 0 0
S SKP 8
ID FL7AACGL0059
KNApSAcK_ID C00006830
NAME Cyanidin 3-(6''-malonylglucoside-3'-(6'''-caffeylglucoside)
CAS_RN 150447-30-6
FORMULA C39H39O22
EXACTMASS 859.193297932
AVERAGEMASS 859.7137600000001
SMILES c(c13)c(O)cc(c1cc(c(c(c4)cc(OC(C(O)6)OC(C(O)C(O)6)COC(=O)C=Cc(c5)cc(O)c(O)c5)c(O)c4)[o+1]3)OC(C2O)OC(C(C2O)O)COC(=O)CC(O)=O)O
M END