Mol:FL7AACGA0006
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.5795 1.1501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5795 1.1501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5795 0.5077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5795 0.5077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0232 0.1865 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0232 0.1865 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4669 0.5077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4669 0.5077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4669 1.1501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4669 1.1501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0232 1.4712 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0232 1.4712 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9106 0.1865 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9106 0.1865 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3543 0.5077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3543 0.5077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3543 1.1501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3543 1.1501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9106 1.4712 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 | + | -0.9106 1.4712 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2018 1.4711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2018 1.4711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7688 1.1438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7688 1.1438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3358 1.4711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3358 1.4711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3358 2.1258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3358 2.1258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7688 2.4531 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7688 2.4531 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2018 2.1258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2018 2.1258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1356 1.4711 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1356 1.4711 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9026 2.4531 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9026 2.4531 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0232 -0.4556 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0232 -0.4556 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0618 -0.2130 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0618 -0.2130 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7688 3.1076 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7688 3.1076 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9547 0.0423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9547 0.0423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3564 -0.4879 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3564 -0.4879 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9349 -0.2630 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9349 -0.2630 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4930 -0.2569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4930 -0.2569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0874 0.1488 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0874 0.1488 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5814 -0.1183 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5814 -0.1183 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7302 -0.5184 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7302 -0.5184 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2663 -0.8193 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2663 -0.8193 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8303 -0.8412 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8303 -0.8412 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7302 -1.2035 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7302 -1.2035 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1665 -1.5290 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1665 -1.5290 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3232 -1.5459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3232 -1.5459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3232 -2.1709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3232 -2.1709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8645 -2.4834 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8645 -2.4834 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4057 -2.1709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4057 -2.1709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4057 -1.5459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4057 -1.5459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8645 -1.2334 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8645 -1.2334 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8645 -3.1076 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8645 -3.1076 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9464 -2.4830 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9464 -2.4830 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9464 -1.2338 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9464 -1.2338 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4211 0.1276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4211 0.1276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1356 -0.2849 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1356 -0.2849 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 1 17 1 0 0 0 0 | + | 1 17 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 20 8 1 0 0 0 0 | + | 20 8 1 0 0 0 0 |
| − | 15 21 1 0 0 0 0 | + | 15 21 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 23 28 1 0 0 0 0 | + | 23 28 1 0 0 0 0 |
| − | 24 29 1 0 0 0 0 | + | 24 29 1 0 0 0 0 |
| − | 25 30 1 0 0 0 0 | + | 25 30 1 0 0 0 0 |
| − | 20 22 1 0 0 0 0 | + | 20 22 1 0 0 0 0 |
| − | 28 31 1 0 0 0 0 | + | 28 31 1 0 0 0 0 |
| − | 31 32 2 0 0 0 0 | + | 31 32 2 0 0 0 0 |
| − | 31 33 1 0 0 0 0 | + | 31 33 1 0 0 0 0 |
| − | 33 34 2 0 0 0 0 | + | 33 34 2 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 2 0 0 0 0 | + | 35 36 2 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 2 0 0 0 0 | + | 37 38 2 0 0 0 0 |
| − | 38 33 1 0 0 0 0 | + | 38 33 1 0 0 0 0 |
| − | 35 39 1 0 0 0 0 | + | 35 39 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 37 41 1 0 0 0 0 | + | 37 41 1 0 0 0 0 |
| − | 26 42 1 0 0 0 0 | + | 26 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 42 43 | + | M SAL 1 2 42 43 |
| − | M SBL 1 1 46 | + | M SBL 1 1 46 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 46 -8.7179 5.0683 | + | M SBV 1 46 -8.7179 5.0683 |
| − | S SKP 8 | + | S SKP 8 |
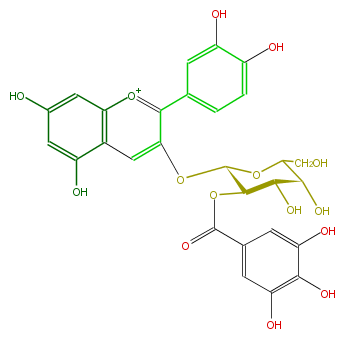
| − | ID FL7AACGA0006 | + | ID FL7AACGA0006 |
| − | KNApSAcK_ID C00006791 | + | KNApSAcK_ID C00006791 |
| − | NAME Cyanidin 3-(2''-galloylgalactoside) | + | NAME Cyanidin 3-(2''-galloylgalactoside) |
| − | CAS_RN 142609-12-9 | + | CAS_RN 142609-12-9 |
| − | FORMULA C28H25O15 | + | FORMULA C28H25O15 |
| − | EXACTMASS 601.11934513 | + | EXACTMASS 601.11934513 |
| − | AVERAGEMASS 601.4891 | + | AVERAGEMASS 601.4891 |
| − | SMILES c(c(O)5)c(cc(O)c(O)5)C(=O)OC(C(O)1)C(Oc(c4)c([o+1]c(c43)cc(cc3O)O)c(c2)ccc(c2O)O)OC(C1O)CO | + | SMILES c(c(O)5)c(cc(O)c(O)5)C(=O)OC(C(O)1)C(Oc(c4)c([o+1]c(c43)cc(cc3O)O)c(c2)ccc(c2O)O)OC(C1O)CO |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-2.5795 1.1501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5795 0.5077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0232 0.1865 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4669 0.5077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4669 1.1501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0232 1.4712 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9106 0.1865 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3543 0.5077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3543 1.1501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9106 1.4712 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
0.2018 1.4711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7688 1.1438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3358 1.4711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3358 2.1258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7688 2.4531 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2018 2.1258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1356 1.4711 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9026 2.4531 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0232 -0.4556 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0618 -0.2130 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7688 3.1076 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9547 0.0423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3564 -0.4879 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9349 -0.2630 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4930 -0.2569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0874 0.1488 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5814 -0.1183 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7302 -0.5184 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2663 -0.8193 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8303 -0.8412 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7302 -1.2035 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1665 -1.5290 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3232 -1.5459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3232 -2.1709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8645 -2.4834 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4057 -2.1709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4057 -1.5459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8645 -1.2334 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8645 -3.1076 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9464 -2.4830 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9464 -1.2338 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4211 0.1276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1356 -0.2849 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
1 17 1 0 0 0 0
14 18 1 0 0 0 0
3 19 1 0 0 0 0
20 8 1 0 0 0 0
15 21 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
23 28 1 0 0 0 0
24 29 1 0 0 0 0
25 30 1 0 0 0 0
20 22 1 0 0 0 0
28 31 1 0 0 0 0
31 32 2 0 0 0 0
31 33 1 0 0 0 0
33 34 2 0 0 0 0
34 35 1 0 0 0 0
35 36 2 0 0 0 0
36 37 1 0 0 0 0
37 38 2 0 0 0 0
38 33 1 0 0 0 0
35 39 1 0 0 0 0
36 40 1 0 0 0 0
37 41 1 0 0 0 0
26 42 1 0 0 0 0
42 43 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 42 43
M SBL 1 1 46
M SMT 1 CH2OH
M SBV 1 46 -8.7179 5.0683
S SKP 8
ID FL7AACGA0006
KNApSAcK_ID C00006791
NAME Cyanidin 3-(2''-galloylgalactoside)
CAS_RN 142609-12-9
FORMULA C28H25O15
EXACTMASS 601.11934513
AVERAGEMASS 601.4891
SMILES c(c(O)5)c(cc(O)c(O)5)C(=O)OC(C(O)1)C(Oc(c4)c([o+1]c(c43)cc(cc3O)O)c(c2)ccc(c2O)O)OC(C1O)CO
M END