Mol:FL5FGGNS0012
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 32 34 0 0 0 0 0 0 0 0999 V2000 | + | 32 34 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.2410 -0.2105 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2410 -0.2105 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2410 -0.8529 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2410 -0.8529 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6847 -1.1741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6847 -1.1741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1284 -0.8529 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1284 -0.8529 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1284 -0.2105 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1284 -0.2105 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6847 0.1107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6847 0.1107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5721 -1.1741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5721 -1.1741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0158 -0.8529 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0158 -0.8529 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0158 -0.2105 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0158 -0.2105 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5721 0.1107 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5721 0.1107 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5721 -1.6749 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5721 -1.6749 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5403 0.1106 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5403 0.1106 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1072 -0.2168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1072 -0.2168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6742 0.1106 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6742 0.1106 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6742 0.7652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6742 0.7652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1072 1.0926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1072 1.0926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5403 0.7652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5403 0.7652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1072 1.7471 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1072 1.7471 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5403 -0.3894 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5403 -0.3894 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4063 -0.8893 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4063 -0.8893 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6847 -2.1741 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6847 -2.1741 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6847 -3.1741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6847 -3.1741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8502 -1.3529 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8502 -1.3529 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7162 -1.8529 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7162 -1.8529 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2410 1.0925 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2410 1.0925 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9555 0.6800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9555 0.6800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5983 0.4082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5983 0.4082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0984 1.2742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0984 1.2742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9555 -0.7614 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9555 -0.7614 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9474 -0.6344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9474 -0.6344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4067 0.6837 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4067 0.6837 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9702 1.5834 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9702 1.5834 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 16 18 1 0 0 0 0 | + | 16 18 1 0 0 0 0 |
| − | 14 19 1 0 0 0 0 | + | 14 19 1 0 0 0 0 |
| − | 19 20 1 0 0 0 0 | + | 19 20 1 0 0 0 0 |
| − | 3 21 1 0 0 0 0 | + | 3 21 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 8 23 1 0 0 0 0 | + | 8 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 15 25 1 0 0 0 0 | + | 15 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 1 27 1 0 0 0 0 | + | 1 27 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 2 29 1 0 0 0 0 | + | 2 29 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 6 31 1 0 0 0 0 | + | 6 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | M STY 1 7 SUP | + | M STY 1 7 SUP |
| − | M SLB 1 7 7 | + | M SLB 1 7 7 |
| − | M SAL 7 2 31 32 | + | M SAL 7 2 31 32 |
| − | M SBL 7 1 33 | + | M SBL 7 1 33 |
| − | M SMT 7 OCH3 | + | M SMT 7 OCH3 |
| − | M SVB 7 33 -1.4067 0.6837 | + | M SVB 7 33 -1.4067 0.6837 |
| − | M STY 1 6 SUP | + | M STY 1 6 SUP |
| − | M SLB 1 6 6 | + | M SLB 1 6 6 |
| − | M SAL 6 2 29 30 | + | M SAL 6 2 29 30 |
| − | M SBL 6 1 31 | + | M SBL 6 1 31 |
| − | M SMT 6 OCH3 | + | M SMT 6 OCH3 |
| − | M SVB 6 31 -2.9555 -0.7614 | + | M SVB 6 31 -2.9555 -0.7614 |
| − | M STY 1 5 SUP | + | M STY 1 5 SUP |
| − | M SLB 1 5 5 | + | M SLB 1 5 5 |
| − | M SAL 5 2 27 28 | + | M SAL 5 2 27 28 |
| − | M SBL 5 1 29 | + | M SBL 5 1 29 |
| − | M SMT 5 OCH3 | + | M SMT 5 OCH3 |
| − | M SVB 5 29 -2.5983 0.4082 | + | M SVB 5 29 -2.5983 0.4082 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 25 26 | + | M SAL 4 2 25 26 |
| − | M SBL 4 1 27 | + | M SBL 4 1 27 |
| − | M SMT 4 OCH3 | + | M SMT 4 OCH3 |
| − | M SVB 4 27 2.241 1.0925 | + | M SVB 4 27 2.241 1.0925 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 23 24 | + | M SAL 3 2 23 24 |
| − | M SBL 3 1 25 | + | M SBL 3 1 25 |
| − | M SMT 3 OCH3 | + | M SMT 3 OCH3 |
| − | M SVB 3 25 0.183 -1.0591 | + | M SVB 3 25 0.183 -1.0591 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 21 22 | + | M SAL 2 2 21 22 |
| − | M SBL 2 1 23 | + | M SBL 2 1 23 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 23 -2.1212 -1.3346 | + | M SVB 2 23 -2.1212 -1.3346 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 19 20 | + | M SAL 1 2 19 20 |
| − | M SBL 1 1 21 | + | M SBL 1 1 21 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 21 1.8838 -0.0957 | + | M SVB 1 21 1.8838 -0.0957 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FGGNS0012 | + | ID FL5FGGNS0012 |
| − | KNApSAcK_ID C00004869 | + | KNApSAcK_ID C00004869 |
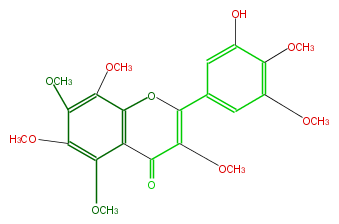
| − | NAME 3'-Hydroxy-3,5,6,7,8,4',5'-heptamethoxyflavone | + | NAME 3'-Hydroxy-3,5,6,7,8,4',5'-heptamethoxyflavone |
| − | CAS_RN 5244-28-0 | + | CAS_RN 5244-28-0 |
| − | FORMULA C22H24O10 | + | FORMULA C22H24O10 |
| − | EXACTMASS 448.136946988 | + | EXACTMASS 448.136946988 |
| − | AVERAGEMASS 448.41996000000006 | + | AVERAGEMASS 448.41996000000006 |
| − | SMILES COC(=C(c(c3)cc(O)c(OC)c3OC)2)C(=O)c(c(OC)1)c(O2)c(OC)c(OC)c1OC | + | SMILES COC(=C(c(c3)cc(O)c(OC)c3OC)2)C(=O)c(c(OC)1)c(O2)c(OC)c(OC)c1OC |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
32 34 0 0 0 0 0 0 0 0999 V2000
-2.2410 -0.2105 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2410 -0.8529 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6847 -1.1741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1284 -0.8529 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1284 -0.2105 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6847 0.1107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5721 -1.1741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0158 -0.8529 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0158 -0.2105 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5721 0.1107 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5721 -1.6749 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5403 0.1106 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1072 -0.2168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6742 0.1106 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6742 0.7652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1072 1.0926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5403 0.7652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1072 1.7471 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5403 -0.3894 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4063 -0.8893 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6847 -2.1741 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6847 -3.1741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8502 -1.3529 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7162 -1.8529 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2410 1.0925 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9555 0.6800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5983 0.4082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0984 1.2742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9555 -0.7614 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9474 -0.6344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4067 0.6837 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9702 1.5834 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
16 18 1 0 0 0 0
14 19 1 0 0 0 0
19 20 1 0 0 0 0
3 21 1 0 0 0 0
21 22 1 0 0 0 0
8 23 1 0 0 0 0
23 24 1 0 0 0 0
15 25 1 0 0 0 0
25 26 1 0 0 0 0
1 27 1 0 0 0 0
27 28 1 0 0 0 0
2 29 1 0 0 0 0
29 30 1 0 0 0 0
6 31 1 0 0 0 0
31 32 1 0 0 0 0
M STY 1 7 SUP
M SLB 1 7 7
M SAL 7 2 31 32
M SBL 7 1 33
M SMT 7 OCH3
M SVB 7 33 -1.4067 0.6837
M STY 1 6 SUP
M SLB 1 6 6
M SAL 6 2 29 30
M SBL 6 1 31
M SMT 6 OCH3
M SVB 6 31 -2.9555 -0.7614
M STY 1 5 SUP
M SLB 1 5 5
M SAL 5 2 27 28
M SBL 5 1 29
M SMT 5 OCH3
M SVB 5 29 -2.5983 0.4082
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 25 26
M SBL 4 1 27
M SMT 4 OCH3
M SVB 4 27 2.241 1.0925
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 23 24
M SBL 3 1 25
M SMT 3 OCH3
M SVB 3 25 0.183 -1.0591
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 21 22
M SBL 2 1 23
M SMT 2 OCH3
M SVB 2 23 -2.1212 -1.3346
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 19 20
M SBL 1 1 21
M SMT 1 OCH3
M SVB 1 21 1.8838 -0.0957
S SKP 8
ID FL5FGGNS0012
KNApSAcK_ID C00004869
NAME 3'-Hydroxy-3,5,6,7,8,4',5'-heptamethoxyflavone
CAS_RN 5244-28-0
FORMULA C22H24O10
EXACTMASS 448.136946988
AVERAGEMASS 448.41996000000006
SMILES COC(=C(c(c3)cc(O)c(OC)c3OC)2)C(=O)c(c(OC)1)c(O2)c(OC)c(OC)c1OC
M END