Mol:FL5FGGGS0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 40 43 0 0 0 0 0 0 0 0999 V2000 | + | 40 43 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.3100 -0.1760 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3100 -0.1760 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3100 -0.8183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3100 -0.8183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7537 -1.1395 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7537 -1.1395 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1974 -0.8183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1974 -0.8183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1974 -0.1760 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1974 -0.1760 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7537 0.1452 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7537 0.1452 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6411 -1.1395 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6411 -1.1395 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0848 -0.8183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0848 -0.8183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0848 -0.1760 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0848 -0.1760 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6411 0.1452 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6411 0.1452 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6411 -1.6403 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6411 -1.6403 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5287 0.1451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5287 0.1451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0382 -0.1822 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0382 -0.1822 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6052 0.1451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6052 0.1451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6052 0.7998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6052 0.7998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0382 1.1271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0382 1.1271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5287 0.7998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5287 0.7998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7537 -1.7816 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7537 -1.7816 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8661 0.1451 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8661 0.1451 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0382 1.7816 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0382 1.7816 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1720 -0.1822 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1720 -0.1822 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0974 0.2254 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.0974 0.2254 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.7966 -0.2956 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.7966 -0.2956 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.3751 -0.1303 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3751 -0.1303 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9571 -0.2956 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.9571 -0.2956 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.2580 0.2254 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.2580 0.2254 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.6794 0.0601 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.6794 0.0601 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.5387 0.0757 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5387 0.0757 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7209 0.5347 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7209 0.5347 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6920 -0.0252 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6920 -0.0252 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4757 0.7182 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4757 0.7182 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0392 1.6179 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0392 1.6179 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1894 1.1187 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1894 1.1187 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9038 0.7062 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9038 0.7062 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9978 -0.6042 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9978 -0.6042 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7122 -1.0169 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7122 -1.0169 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2188 -1.3183 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2188 -1.3183 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6473 -1.8182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6473 -1.8182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1567 -0.5879 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1567 -0.5879 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1567 -1.4129 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1567 -1.4129 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 16 20 1 0 0 0 0 | + | 16 20 1 0 0 0 0 |
| − | 14 21 1 0 0 0 0 | + | 14 21 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 23 21 1 0 0 0 0 | + | 23 21 1 0 0 0 0 |
| − | 6 31 1 0 0 0 0 | + | 6 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 15 33 1 0 0 0 0 | + | 15 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 2 35 1 0 0 0 0 | + | 2 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 8 37 1 0 0 0 0 | + | 8 37 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 25 39 1 0 0 0 0 | + | 25 39 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | M STY 1 5 SUP | + | M STY 1 5 SUP |
| − | M SLB 1 5 5 | + | M SLB 1 5 5 |
| − | M SAL 5 2 39 40 | + | M SAL 5 2 39 40 |
| − | M SBL 5 1 42 | + | M SBL 5 1 42 |
| − | M SMT 5 CH2OH | + | M SMT 5 CH2OH |
| − | M SVB 5 42 3.31 -0.2689 | + | M SVB 5 42 3.31 -0.2689 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 37 38 | + | M SAL 4 2 37 38 |
| − | M SBL 4 1 40 | + | M SBL 4 1 40 |
| − | M SMT 4 OCH3 | + | M SMT 4 OCH3 |
| − | M SVB 4 40 -0.886 -1.0246 | + | M SVB 4 40 -0.886 -1.0246 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 35 36 | + | M SAL 3 2 35 36 |
| − | M SBL 3 1 38 | + | M SBL 3 1 38 |
| − | M SMT 3 OCH3 | + | M SMT 3 OCH3 |
| − | M SVB 3 38 -4.0245 -0.7269 | + | M SVB 3 38 -4.0245 -0.7269 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 33 34 | + | M SAL 2 2 33 34 |
| − | M SBL 2 1 36 | + | M SBL 2 1 36 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 36 1.1894 1.1187 | + | M SVB 2 36 1.1894 1.1187 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 31 32 | + | M SAL 1 2 31 32 |
| − | M SBL 1 1 34 | + | M SBL 1 1 34 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 34 -2.4757 0.7182 | + | M SVB 1 34 -2.4757 0.7182 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FGGGS0001 | + | ID FL5FGGGS0001 |
| − | KNApSAcK_ID C00005797 | + | KNApSAcK_ID C00005797 |
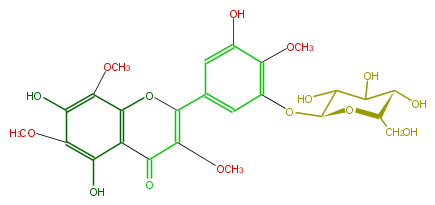
| − | NAME 5,7,3',5'-Tetrahydroxy-3,6,8,4'-tetramethoxyflavone 3'-glucoside | + | NAME 5,7,3',5'-Tetrahydroxy-3,6,8,4'-tetramethoxyflavone 3'-glucoside |
| − | CAS_RN 101021-27-6 | + | CAS_RN 101021-27-6 |
| − | FORMULA C25H28O15 | + | FORMULA C25H28O15 |
| − | EXACTMASS 568.1428202259999 | + | EXACTMASS 568.1428202259999 |
| − | AVERAGEMASS 568.48082 | + | AVERAGEMASS 568.48082 |
| − | SMILES c(OC)(c4O)c(O)c(OC)c(c34)OC(=C(OC)C3=O)c(c1)cc(c(c1O[C@@H](C2O)O[C@@H]([C@H](O)C2O)CO)OC)O | + | SMILES c(OC)(c4O)c(O)c(OC)c(c34)OC(=C(OC)C3=O)c(c1)cc(c(c1O[C@@H](C2O)O[C@@H]([C@H](O)C2O)CO)OC)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
40 43 0 0 0 0 0 0 0 0999 V2000
-3.3100 -0.1760 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3100 -0.8183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7537 -1.1395 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1974 -0.8183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1974 -0.1760 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7537 0.1452 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6411 -1.1395 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0848 -0.8183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0848 -0.1760 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6411 0.1452 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6411 -1.6403 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5287 0.1451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0382 -0.1822 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6052 0.1451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6052 0.7998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0382 1.1271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5287 0.7998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7537 -1.7816 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8661 0.1451 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0382 1.7816 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1720 -0.1822 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0974 0.2254 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.7966 -0.2956 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.3751 -0.1303 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9571 -0.2956 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.2580 0.2254 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.6794 0.0601 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.5387 0.0757 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7209 0.5347 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6920 -0.0252 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4757 0.7182 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0392 1.6179 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1894 1.1187 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9038 0.7062 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9978 -0.6042 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.7122 -1.0169 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2188 -1.3183 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6473 -1.8182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1567 -0.5879 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1567 -1.4129 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
16 20 1 0 0 0 0
14 21 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
23 21 1 0 0 0 0
6 31 1 0 0 0 0
31 32 1 0 0 0 0
15 33 1 0 0 0 0
33 34 1 0 0 0 0
2 35 1 0 0 0 0
35 36 1 0 0 0 0
8 37 1 0 0 0 0
37 38 1 0 0 0 0
25 39 1 0 0 0 0
39 40 1 0 0 0 0
M STY 1 5 SUP
M SLB 1 5 5
M SAL 5 2 39 40
M SBL 5 1 42
M SMT 5 CH2OH
M SVB 5 42 3.31 -0.2689
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 37 38
M SBL 4 1 40
M SMT 4 OCH3
M SVB 4 40 -0.886 -1.0246
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 35 36
M SBL 3 1 38
M SMT 3 OCH3
M SVB 3 38 -4.0245 -0.7269
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 33 34
M SBL 2 1 36
M SMT 2 OCH3
M SVB 2 36 1.1894 1.1187
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 31 32
M SBL 1 1 34
M SMT 1 OCH3
M SVB 1 34 -2.4757 0.7182
S SKP 8
ID FL5FGGGS0001
KNApSAcK_ID C00005797
NAME 5,7,3',5'-Tetrahydroxy-3,6,8,4'-tetramethoxyflavone 3'-glucoside
CAS_RN 101021-27-6
FORMULA C25H28O15
EXACTMASS 568.1428202259999
AVERAGEMASS 568.48082
SMILES c(OC)(c4O)c(O)c(OC)c(c34)OC(=C(OC)C3=O)c(c1)cc(c(c1O[C@@H](C2O)O[C@@H]([C@H](O)C2O)CO)OC)O
M END