Mol:FL5FEAGL0008
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 36 39 0 0 0 0 0 0 0 0999 V2000 | + | 36 39 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.4013 -4.2281 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4013 -4.2281 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7809 -4.3944 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7809 -4.3944 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3267 -3.9402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3267 -3.9402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4930 -3.3197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4930 -3.3197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1134 -3.1534 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1134 -3.1534 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5676 -3.6076 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5676 -3.6076 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0387 -2.8655 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0387 -2.8655 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2050 -2.2449 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2050 -2.2449 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8254 -2.0787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8254 -2.0787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2797 -2.5330 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2797 -2.5330 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4450 -2.9950 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4450 -2.9950 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9917 -1.4585 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9917 -1.4585 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5287 -0.9955 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5287 -0.9955 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6982 -0.3631 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6982 -0.3631 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3305 -0.1937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3305 -0.1937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7934 -0.6566 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7934 -0.6566 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6240 -1.2890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6240 -1.2890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4943 -1.7613 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4943 -1.7613 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2261 -0.5012 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.2261 -0.5012 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.8384 -1.1726 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.8384 -1.1726 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.5839 -0.9596 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5839 -0.9596 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3339 -1.1726 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.3339 -1.1726 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.7216 -0.5012 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.7216 -0.5012 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.9761 -0.7142 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.9761 -0.7142 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.4990 -0.6960 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4990 -0.6960 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2781 -0.1911 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2781 -0.1911 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4071 -0.6849 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4071 -0.6849 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4999 0.4385 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4999 0.4385 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0541 -5.0608 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0541 -5.0608 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4334 -5.9860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4334 -5.9860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0915 -4.4130 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0915 -4.4130 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0575 -4.6717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0575 -4.6717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6392 -4.1988 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6392 -4.1988 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6051 -4.4577 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6051 -4.4577 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1606 -1.5031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1606 -1.5031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3034 -2.4929 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3034 -2.4929 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 19 20 1 0 0 0 0 | + | 19 20 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 19 25 1 0 0 0 0 | + | 19 25 1 0 0 0 0 |
| − | 24 26 1 0 0 0 0 | + | 24 26 1 0 0 0 0 |
| − | 23 27 1 0 0 0 0 | + | 23 27 1 0 0 0 0 |
| − | 20 18 1 0 0 0 0 | + | 20 18 1 0 0 0 0 |
| − | 18 8 1 0 0 0 0 | + | 18 8 1 0 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 15 28 1 0 0 0 0 | + | 15 28 1 0 0 0 0 |
| − | 2 29 1 0 0 0 0 | + | 2 29 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 1 31 1 0 0 0 0 | + | 1 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 3 33 1 0 0 0 0 | + | 3 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 22 35 1 0 0 0 0 | + | 22 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 35 36 | + | M SAL 4 2 35 36 |
| − | M SBL 4 1 38 | + | M SBL 4 1 38 |
| − | M SMT 4 CH2OH | + | M SMT 4 CH2OH |
| − | M SVB 4 38 3.1606 -1.5031 | + | M SVB 4 38 3.1606 -1.5031 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 33 34 | + | M SAL 3 2 33 34 |
| − | M SBL 3 1 36 | + | M SBL 3 1 36 |
| − | M SMT 3 OCH3 | + | M SMT 3 OCH3 |
| − | M SVB 3 36 -2.5728 -0.924 | + | M SVB 3 36 -2.5728 -0.924 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 31 32 | + | M SAL 2 2 31 32 |
| − | M SBL 2 1 34 | + | M SBL 2 1 34 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 34 -3.0499 0.8188 | + | M SVB 2 34 -3.0499 0.8188 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 29 30 | + | M SAL 1 2 29 30 |
| − | M SBL 1 1 32 | + | M SBL 1 1 32 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 32 -3.4071 -0.3509 | + | M SVB 1 32 -3.4071 -0.3509 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FEAGL0008 | + | ID FL5FEAGL0008 |
| − | KNApSAcK_ID C00005333 | + | KNApSAcK_ID C00005333 |
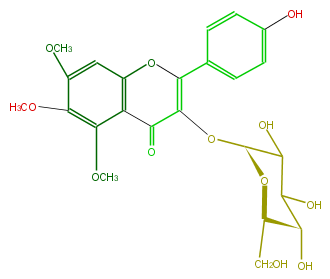
| − | NAME Candidol 3-glucoside | + | NAME Candidol 3-glucoside |
| − | CAS_RN 141304-80-5 | + | CAS_RN 141304-80-5 |
| − | FORMULA C24H26O12 | + | FORMULA C24H26O12 |
| − | EXACTMASS 506.142426296 | + | EXACTMASS 506.142426296 |
| − | AVERAGEMASS 506.45604 | + | AVERAGEMASS 506.45604 |
| − | SMILES OC([C@@H]1OC(=C3c(c4)ccc(c4)O)C(c(c2O3)c(OC)c(OC)c(c2)OC)=O)C([C@@H](O)[C@@H](CO)O1)O | + | SMILES OC([C@@H]1OC(=C3c(c4)ccc(c4)O)C(c(c2O3)c(OC)c(OC)c(c2)OC)=O)C([C@@H](O)[C@@H](CO)O1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
36 39 0 0 0 0 0 0 0 0999 V2000
-1.4013 -4.2281 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7809 -4.3944 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3267 -3.9402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4930 -3.3197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1134 -3.1534 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5676 -3.6076 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0387 -2.8655 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2050 -2.2449 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8254 -2.0787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2797 -2.5330 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4450 -2.9950 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9917 -1.4585 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5287 -0.9955 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6982 -0.3631 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3305 -0.1937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7934 -0.6566 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6240 -1.2890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4943 -1.7613 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2261 -0.5012 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.8384 -1.1726 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.5839 -0.9596 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3339 -1.1726 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.7216 -0.5012 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.9761 -0.7142 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.4990 -0.6960 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2781 -0.1911 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4071 -0.6849 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4999 0.4385 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0541 -5.0608 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4334 -5.9860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0915 -4.4130 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0575 -4.6717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6392 -4.1988 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6051 -4.4577 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1606 -1.5031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3034 -2.4929 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
19 20 1 0 0 0 0
20 21 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 19 1 0 0 0 0
19 25 1 0 0 0 0
24 26 1 0 0 0 0
23 27 1 0 0 0 0
20 18 1 0 0 0 0
18 8 1 0 0 0 0
22 21 1 1 0 0 0
15 28 1 0 0 0 0
2 29 1 0 0 0 0
29 30 1 0 0 0 0
1 31 1 0 0 0 0
31 32 1 0 0 0 0
3 33 1 0 0 0 0
33 34 1 0 0 0 0
22 35 1 0 0 0 0
35 36 1 0 0 0 0
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 35 36
M SBL 4 1 38
M SMT 4 CH2OH
M SVB 4 38 3.1606 -1.5031
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 33 34
M SBL 3 1 36
M SMT 3 OCH3
M SVB 3 36 -2.5728 -0.924
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 31 32
M SBL 2 1 34
M SMT 2 OCH3
M SVB 2 34 -3.0499 0.8188
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 29 30
M SBL 1 1 32
M SMT 1 OCH3
M SVB 1 32 -3.4071 -0.3509
S SKP 8
ID FL5FEAGL0008
KNApSAcK_ID C00005333
NAME Candidol 3-glucoside
CAS_RN 141304-80-5
FORMULA C24H26O12
EXACTMASS 506.142426296
AVERAGEMASS 506.45604
SMILES OC([C@@H]1OC(=C3c(c4)ccc(c4)O)C(c(c2O3)c(OC)c(OC)c(c2)OC)=O)C([C@@H](O)[C@@H](CO)O1)O
M END