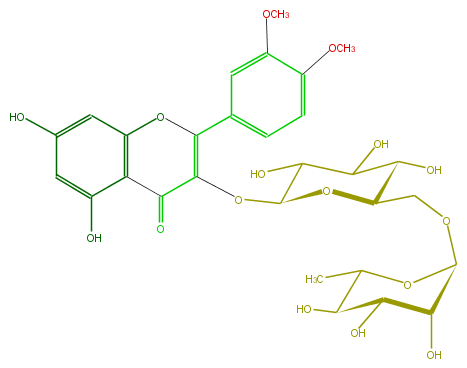
Mol:FL5FAFGL0002
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 45 49 0 0 0 0 0 0 0 0999 V2000 | + | 45 49 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.6578 0.3839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6578 0.3839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6578 -0.4255 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6578 -0.4255 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9568 -0.8302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9568 -0.8302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2558 -0.4255 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2558 -0.4255 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2558 0.3839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2558 0.3839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9568 0.7887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9568 0.7887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5548 -0.8302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5548 -0.8302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8537 -0.4255 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8537 -0.4255 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8537 0.3839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8537 0.3839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5548 0.7887 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5548 0.7887 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5548 -1.4613 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5548 -1.4613 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1530 0.7886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1530 0.7886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5615 0.3761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5615 0.3761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2760 0.7886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2760 0.7886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2760 1.6136 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2760 1.6136 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5615 2.0260 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5615 2.0260 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1530 1.6136 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1530 1.6136 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9568 -1.6393 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9568 -1.6393 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4455 -2.3113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4455 -2.3113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9569 -3.1574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9569 -3.1574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8964 -2.8890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8964 -2.8890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8415 -3.1574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8415 -3.1574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3586 -2.1873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3586 -2.1873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3906 -2.5797 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3906 -2.5797 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6263 -2.4657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6263 -2.4657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2764 -0.1621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2764 -0.1621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8292 -0.9669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8292 -0.9669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7672 -0.6970 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7672 -0.6970 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7122 -0.9655 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7122 -0.9655 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2009 -0.1194 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2009 -0.1194 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2615 -0.3877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2615 -0.3877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0090 -0.8810 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0090 -0.8810 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7733 0.2396 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7733 0.2396 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8327 -0.2845 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8327 -0.2845 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5062 -0.8019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5062 -0.8019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1562 -1.3008 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1562 -1.3008 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3799 -3.0668 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3799 -3.0668 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3251 -3.5440 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3251 -3.5440 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8415 -3.9818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8415 -3.9818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4562 -0.3557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4562 -0.3557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3586 0.7886 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3586 0.7886 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8857 2.1694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8857 2.1694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9771 2.7995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9771 2.7995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5613 2.8510 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5613 2.8510 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1174 3.9818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1174 3.9818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 20 1 0 0 0 0 | + | 19 20 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 19 25 1 0 0 0 0 | + | 19 25 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 1 0 0 0 | + | 27 28 1 1 0 0 0 |
| − | 28 29 1 1 0 0 0 | + | 28 29 1 1 0 0 0 |
| − | 30 29 1 1 0 0 0 | + | 30 29 1 1 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 26 1 0 0 0 0 | + | 31 26 1 0 0 0 0 |
| − | 31 33 1 0 0 0 0 | + | 31 33 1 0 0 0 0 |
| − | 30 34 1 0 0 0 0 | + | 30 34 1 0 0 0 0 |
| − | 29 35 1 0 0 0 0 | + | 29 35 1 0 0 0 0 |
| − | 23 36 1 0 0 0 0 | + | 23 36 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 20 37 1 0 0 0 0 | + | 20 37 1 0 0 0 0 |
| − | 21 38 1 0 0 0 0 | + | 21 38 1 0 0 0 0 |
| − | 22 39 1 0 0 0 0 | + | 22 39 1 0 0 0 0 |
| − | 32 8 1 0 0 0 0 | + | 32 8 1 0 0 0 0 |
| − | 27 32 1 0 0 0 0 | + | 27 32 1 0 0 0 0 |
| − | 26 40 1 0 0 0 0 | + | 26 40 1 0 0 0 0 |
| − | 1 41 1 0 0 0 0 | + | 1 41 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 15 42 1 0 0 0 0 | + | 15 42 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 16 44 1 0 0 0 0 | + | 16 44 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 42 43 | + | M SAL 1 2 42 43 |
| − | M SBL 1 1 47 | + | M SBL 1 1 47 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 47 -0.6097 -0.5558 | + | M SBV 1 47 -0.6097 -0.5558 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 44 45 | + | M SAL 2 2 44 45 |
| − | M SBL 2 1 49 | + | M SBL 2 1 49 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SBV 2 49 0.0002 -0.8250 | + | M SBV 2 49 0.0002 -0.8250 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FAFGL0002 | + | ID FL5FAFGL0002 |
| − | FORMULA C29H34O16 | + | FORMULA C29H34O16 |
| − | EXACTMASS 638.18468504 | + | EXACTMASS 638.18468504 |
| − | AVERAGEMASS 638.57066 | + | AVERAGEMASS 638.57066 |
| − | SMILES O(C)c(c1)c(ccc1C(=C(OC(O4)C(O)C(O)C(C4COC(O5)C(C(C(O)C(C)5)O)O)O)3)Oc(c(C(=O)3)2)cc(O)cc2O)OC | + | SMILES O(C)c(c1)c(ccc1C(=C(OC(O4)C(O)C(O)C(C4COC(O5)C(C(C(O)C(C)5)O)O)O)3)Oc(c(C(=O)3)2)cc(O)cc2O)OC |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
45 49 0 0 0 0 0 0 0 0999 V2000
-3.6578 0.3839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6578 -0.4255 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9568 -0.8302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2558 -0.4255 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2558 0.3839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9568 0.7887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5548 -0.8302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8537 -0.4255 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8537 0.3839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5548 0.7887 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5548 -1.4613 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1530 0.7886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5615 0.3761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2760 0.7886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2760 1.6136 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5615 2.0260 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1530 1.6136 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9568 -1.6393 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4455 -2.3113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9569 -3.1574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8964 -2.8890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8415 -3.1574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3586 -2.1873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3906 -2.5797 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6263 -2.4657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2764 -0.1621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8292 -0.9669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7672 -0.6970 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7122 -0.9655 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2009 -0.1194 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2615 -0.3877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0090 -0.8810 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7733 0.2396 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8327 -0.2845 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5062 -0.8019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1562 -1.3008 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3799 -3.0668 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3251 -3.5440 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8415 -3.9818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4562 -0.3557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3586 0.7886 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8857 2.1694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9771 2.7995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5613 2.8510 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1174 3.9818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 20 1 0 0 0 0
20 21 1 1 0 0 0
21 22 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 19 1 0 0 0 0
19 25 1 0 0 0 0
26 27 1 0 0 0 0
27 28 1 1 0 0 0
28 29 1 1 0 0 0
30 29 1 1 0 0 0
30 31 1 0 0 0 0
31 26 1 0 0 0 0
31 33 1 0 0 0 0
30 34 1 0 0 0 0
29 35 1 0 0 0 0
23 36 1 0 0 0 0
35 36 1 0 0 0 0
20 37 1 0 0 0 0
21 38 1 0 0 0 0
22 39 1 0 0 0 0
32 8 1 0 0 0 0
27 32 1 0 0 0 0
26 40 1 0 0 0 0
1 41 1 0 0 0 0
42 43 1 0 0 0 0
15 42 1 0 0 0 0
44 45 1 0 0 0 0
16 44 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 42 43
M SBL 1 1 47
M SMT 1 OCH3
M SBV 1 47 -0.6097 -0.5558
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 44 45
M SBL 2 1 49
M SMT 2 OCH3
M SBV 2 49 0.0002 -0.8250
S SKP 5
ID FL5FAFGL0002
FORMULA C29H34O16
EXACTMASS 638.18468504
AVERAGEMASS 638.57066
SMILES O(C)c(c1)c(ccc1C(=C(OC(O4)C(O)C(O)C(C4COC(O5)C(C(C(O)C(C)5)O)O)O)3)Oc(c(C(=O)3)2)cc(O)cc2O)OC
M END