Mol:FL5FADGS0014
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.0559 0.1125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0559 0.1125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0559 -0.7125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0559 -0.7125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3414 -1.1250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3414 -1.1250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3731 -0.7125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3731 -0.7125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3731 0.1125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3731 0.1125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3414 0.5250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3414 0.5250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0875 -1.1250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0875 -1.1250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8020 -0.7125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8020 -0.7125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8020 0.1125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8020 0.1125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0875 0.5250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0875 0.5250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0875 -1.7682 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0875 -1.7682 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7016 0.6838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7016 0.6838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4298 0.2633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4298 0.2633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1579 0.6838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1579 0.6838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1579 1.5246 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1579 1.5246 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4298 1.9450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4298 1.9450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7016 1.5246 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7016 1.5246 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3414 -1.9497 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3414 -1.9497 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8590 1.9799 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8590 1.9799 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6824 0.5227 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6824 0.5227 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3722 -1.2827 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3722 -1.2827 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2702 0.6476 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2702 0.6476 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7934 0.0183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7934 0.0183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1070 0.2853 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1070 0.2853 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4445 0.2923 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4445 0.2923 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9259 0.7738 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9259 0.7738 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5264 0.4569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5264 0.4569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8590 0.4755 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8590 0.4755 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5145 0.1457 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5145 0.1457 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4853 -0.2321 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4853 -0.2321 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1442 -2.7033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1442 -2.7033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8318 -3.1003 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8318 -3.1003 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6136 -2.3369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6136 -2.3369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8318 -1.5688 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8318 -1.5688 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1442 -1.1718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1442 -1.1718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3624 -1.9352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3624 -1.9352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4278 -1.9774 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4278 -1.9774 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1373 -3.4141 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1373 -3.4141 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6352 -3.7740 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6352 -3.7740 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0876 -2.7096 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0876 -2.7096 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0671 1.0926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0671 1.0926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7901 1.8697 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7901 1.8697 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4239 2.6216 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4239 2.6216 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9908 3.7740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9908 3.7740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 1 20 1 0 0 0 0 | + | 1 20 1 0 0 0 0 |
| − | 21 8 1 0 0 0 0 | + | 21 8 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 1 0 0 0 | + | 35 36 1 1 0 0 0 |
| − | 36 31 1 1 0 0 0 | + | 36 31 1 1 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 31 38 1 0 0 0 0 | + | 31 38 1 0 0 0 0 |
| − | 32 39 1 0 0 0 0 | + | 32 39 1 0 0 0 0 |
| − | 33 40 1 0 0 0 0 | + | 33 40 1 0 0 0 0 |
| − | 35 21 1 0 0 0 0 | + | 35 21 1 0 0 0 0 |
| − | 27 41 1 0 0 0 0 | + | 27 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 16 43 1 0 0 0 0 | + | 16 43 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 41 42 | + | M SAL 1 2 41 42 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 45 0.5407 -0.6357 | + | M SBV 1 45 0.5407 -0.6357 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 43 44 | + | M SAL 2 2 43 44 |
| − | M SBL 2 1 48 | + | M SBL 2 1 48 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SBV 2 48 0.0059 -0.6766 | + | M SBV 2 48 0.0059 -0.6766 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FADGS0014 | + | ID FL5FADGS0014 |
| − | FORMULA C28H32O16 | + | FORMULA C28H32O16 |
| − | EXACTMASS 624.1690349759999 | + | EXACTMASS 624.1690349759999 |
| − | AVERAGEMASS 624.54408 | + | AVERAGEMASS 624.54408 |
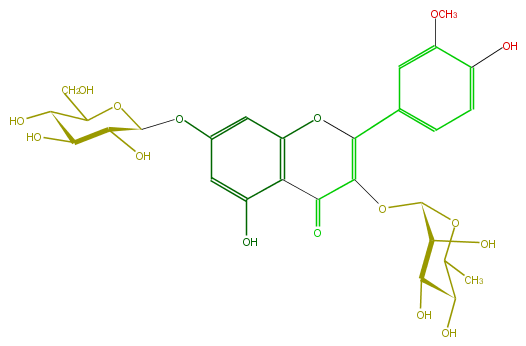
| − | SMILES O(c(c5)cc(O3)c(c5O)C(C(=C(c(c4)cc(OC)c(O)c4)3)OC(C2O)OC(C)C(C(O)2)O)=O)C(O1)C(O)C(O)C(O)C(CO)1 | + | SMILES O(c(c5)cc(O3)c(c5O)C(C(=C(c(c4)cc(OC)c(O)c4)3)OC(C2O)OC(C)C(C(O)2)O)=O)C(O1)C(O)C(O)C(O)C(CO)1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
-1.0559 0.1125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0559 -0.7125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3414 -1.1250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3731 -0.7125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3731 0.1125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3414 0.5250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0875 -1.1250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8020 -0.7125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8020 0.1125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0875 0.5250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0875 -1.7682 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7016 0.6838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4298 0.2633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1579 0.6838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1579 1.5246 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4298 1.9450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7016 1.5246 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3414 -1.9497 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.8590 1.9799 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6824 0.5227 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3722 -1.2827 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2702 0.6476 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7934 0.0183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1070 0.2853 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4445 0.2923 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9259 0.7738 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5264 0.4569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.8590 0.4755 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5145 0.1457 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4853 -0.2321 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1442 -2.7033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8318 -3.1003 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6136 -2.3369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8318 -1.5688 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1442 -1.1718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3624 -1.9352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4278 -1.9774 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1373 -3.4141 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6352 -3.7740 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0876 -2.7096 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0671 1.0926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7901 1.8697 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4239 2.6216 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9908 3.7740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
4 3 1 0 0 0 0
1 20 1 0 0 0 0
21 8 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
25 20 1 0 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 36 1 1 0 0 0
36 31 1 1 0 0 0
36 37 1 0 0 0 0
31 38 1 0 0 0 0
32 39 1 0 0 0 0
33 40 1 0 0 0 0
35 21 1 0 0 0 0
27 41 1 0 0 0 0
41 42 1 0 0 0 0
43 44 1 0 0 0 0
16 43 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 41 42
M SBL 1 1 45
M SMT 1 ^ CH2OH
M SBV 1 45 0.5407 -0.6357
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 43 44
M SBL 2 1 48
M SMT 2 OCH3
M SBV 2 48 0.0059 -0.6766
S SKP 5
ID FL5FADGS0014
FORMULA C28H32O16
EXACTMASS 624.1690349759999
AVERAGEMASS 624.54408
SMILES O(c(c5)cc(O3)c(c5O)C(C(=C(c(c4)cc(OC)c(O)c4)3)OC(C2O)OC(C)C(C(O)2)O)=O)C(O1)C(O)C(O)C(O)C(CO)1
M END