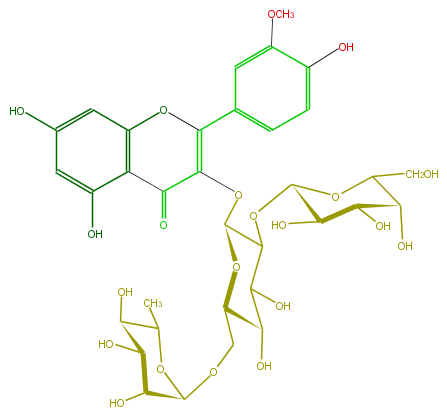
Mol:FL5FADGL0021
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 55 60 0 0 0 0 0 0 0 0999 V2000 | + | 55 60 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.1899 0.9707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1899 0.9707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1899 0.1457 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1899 0.1457 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4755 -0.2668 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4755 -0.2668 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7611 0.1457 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7611 0.1457 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7611 0.9707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7611 0.9707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4755 1.3832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4755 1.3832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0467 -0.2668 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0467 -0.2668 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3322 0.1457 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3322 0.1457 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3322 0.9707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3322 0.9707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0467 1.3832 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0467 1.3832 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0467 -0.9099 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0467 -0.9099 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3819 1.3830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3819 1.3830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1101 0.9626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1101 0.9626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8383 1.3830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8383 1.3830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8383 2.2238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8383 2.2238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1101 2.6442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1101 2.6442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3819 2.2238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3819 2.2238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9041 1.3830 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9041 1.3830 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4641 -0.3139 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4641 -0.3139 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4755 -1.0914 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4755 -1.0914 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5393 2.6583 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5393 2.6583 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9231 -1.3560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9231 -1.3560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1747 -0.9240 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1747 -0.9240 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4121 -1.7549 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4121 -1.7549 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1747 -2.5909 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1747 -2.5909 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9231 -3.0231 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9231 -3.0231 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6857 -2.1921 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6857 -2.1921 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7520 -0.7212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7520 -0.7212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2784 -2.5343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2784 -2.5343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9039 -3.7329 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9039 -3.7329 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3491 -3.3190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3491 -3.3190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4054 -4.2773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4054 -4.2773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6374 -4.4127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6374 -4.4127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0952 -3.7815 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0952 -3.7815 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1517 -2.9994 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1517 -2.9994 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9198 -2.8639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9198 -2.8639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4620 -3.4953 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4620 -3.4953 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8882 -2.2538 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8882 -2.2538 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3693 -2.4652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3693 -2.4652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1283 -3.2833 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1283 -3.2833 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9084 -4.4732 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9084 -4.4732 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0378 -3.8551 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0378 -3.8551 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5153 -0.1141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5153 -0.1141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0834 -0.8641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0834 -0.8641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7997 -0.5472 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7997 -0.5472 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6910 -0.5374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6910 -0.5374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1174 0.0364 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1174 0.0364 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4015 -0.3413 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4015 -0.3413 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3448 -0.9099 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3448 -0.9099 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2778 -0.9265 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2778 -0.9265 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7257 -1.3344 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7257 -1.3344 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8756 0.1206 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8756 0.1206 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9041 1.4044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9041 1.4044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1248 3.3208 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1248 3.3208 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6917 4.4732 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6917 4.4732 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 8 19 1 0 0 0 0 | + | 8 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 21 15 1 0 0 0 0 | + | 21 15 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 25 31 1 0 0 0 0 | + | 25 31 1 0 0 0 0 |
| − | 23 19 1 0 0 0 0 | + | 23 19 1 0 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 1 0 0 0 | + | 36 37 1 1 0 0 0 |
| − | 37 32 1 1 0 0 0 | + | 37 32 1 1 0 0 0 |
| − | 36 38 1 0 0 0 0 | + | 36 38 1 0 0 0 0 |
| − | 35 39 1 0 0 0 0 | + | 35 39 1 0 0 0 0 |
| − | 37 40 1 0 0 0 0 | + | 37 40 1 0 0 0 0 |
| − | 32 41 1 0 0 0 0 | + | 32 41 1 0 0 0 0 |
| − | 33 42 1 0 0 0 0 | + | 33 42 1 0 0 0 0 |
| − | 42 31 1 0 0 0 0 | + | 42 31 1 0 0 0 0 |
| − | 43 44 1 1 0 0 0 | + | 43 44 1 1 0 0 0 |
| − | 44 45 1 1 0 0 0 | + | 44 45 1 1 0 0 0 |
| − | 46 45 1 1 0 0 0 | + | 46 45 1 1 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 48 43 1 0 0 0 0 | + | 48 43 1 0 0 0 0 |
| − | 44 49 1 0 0 0 0 | + | 44 49 1 0 0 0 0 |
| − | 45 50 1 0 0 0 0 | + | 45 50 1 0 0 0 0 |
| − | 43 28 1 0 0 0 0 | + | 43 28 1 0 0 0 0 |
| − | 46 51 1 0 0 0 0 | + | 46 51 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 47 52 1 0 0 0 0 | + | 47 52 1 0 0 0 0 |
| − | 54 55 1 0 0 0 0 | + | 54 55 1 0 0 0 0 |
| − | 16 54 1 0 0 0 0 | + | 16 54 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 52 53 | + | M SAL 1 2 52 53 |
| − | M SBL 1 1 58 | + | M SBL 1 1 58 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 58 -0.7582 -0.0841 | + | M SBV 1 58 -0.7582 -0.0841 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 54 55 | + | M SAL 2 2 54 55 |
| − | M SBL 2 1 60 | + | M SBL 2 1 60 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SBV 2 60 -0.0147 -0.6766 | + | M SBV 2 60 -0.0147 -0.6766 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FADGL0021 | + | ID FL5FADGL0021 |
| − | FORMULA C34H42O21 | + | FORMULA C34H42O21 |
| − | EXACTMASS 786.2218584059999 | + | EXACTMASS 786.2218584059999 |
| − | AVERAGEMASS 786.68468 | + | AVERAGEMASS 786.68468 |
| − | SMILES C(C6O)(C(O)C(OC(C)6)OCC(O1)C(O)C(O)C(OC(O5)C(O)C(C(O)C5CO)O)C(OC(C3=O)=C(c(c4)cc(OC)c(c4)O)Oc(c32)cc(O)cc(O)2)1)O | + | SMILES C(C6O)(C(O)C(OC(C)6)OCC(O1)C(O)C(O)C(OC(O5)C(O)C(C(O)C5CO)O)C(OC(C3=O)=C(c(c4)cc(OC)c(c4)O)Oc(c32)cc(O)cc(O)2)1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
55 60 0 0 0 0 0 0 0 0999 V2000
-3.1899 0.9707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1899 0.1457 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4755 -0.2668 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7611 0.1457 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7611 0.9707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4755 1.3832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0467 -0.2668 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3322 0.1457 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3322 0.9707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0467 1.3832 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0467 -0.9099 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3819 1.3830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1101 0.9626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8383 1.3830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8383 2.2238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1101 2.6442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3819 2.2238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9041 1.3830 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4641 -0.3139 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4755 -1.0914 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5393 2.6583 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9231 -1.3560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1747 -0.9240 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4121 -1.7549 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1747 -2.5909 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9231 -3.0231 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6857 -2.1921 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7520 -0.7212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2784 -2.5343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9039 -3.7329 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3491 -3.3190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4054 -4.2773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6374 -4.4127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0952 -3.7815 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1517 -2.9994 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9198 -2.8639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4620 -3.4953 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8882 -2.2538 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3693 -2.4652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1283 -3.2833 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9084 -4.4732 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0378 -3.8551 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5153 -0.1141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0834 -0.8641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7997 -0.5472 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6910 -0.5374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1174 0.0364 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4015 -0.3413 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3448 -0.9099 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2778 -0.9265 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7257 -1.3344 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8756 0.1206 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9041 1.4044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1248 3.3208 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6917 4.4732 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
8 19 1 0 0 0 0
3 20 1 0 0 0 0
21 15 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
25 31 1 0 0 0 0
23 19 1 0 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 37 1 1 0 0 0
37 32 1 1 0 0 0
36 38 1 0 0 0 0
35 39 1 0 0 0 0
37 40 1 0 0 0 0
32 41 1 0 0 0 0
33 42 1 0 0 0 0
42 31 1 0 0 0 0
43 44 1 1 0 0 0
44 45 1 1 0 0 0
46 45 1 1 0 0 0
46 47 1 0 0 0 0
47 48 1 0 0 0 0
48 43 1 0 0 0 0
44 49 1 0 0 0 0
45 50 1 0 0 0 0
43 28 1 0 0 0 0
46 51 1 0 0 0 0
52 53 1 0 0 0 0
47 52 1 0 0 0 0
54 55 1 0 0 0 0
16 54 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 52 53
M SBL 1 1 58
M SMT 1 CH2OH
M SBV 1 58 -0.7582 -0.0841
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 54 55
M SBL 2 1 60
M SMT 2 OCH3
M SBV 2 60 -0.0147 -0.6766
S SKP 5
ID FL5FADGL0021
FORMULA C34H42O21
EXACTMASS 786.2218584059999
AVERAGEMASS 786.68468
SMILES C(C6O)(C(O)C(OC(C)6)OCC(O1)C(O)C(O)C(OC(O5)C(O)C(C(O)C5CO)O)C(OC(C3=O)=C(c(c4)cc(OC)c(c4)O)Oc(c32)cc(O)cc(O)2)1)O
M END