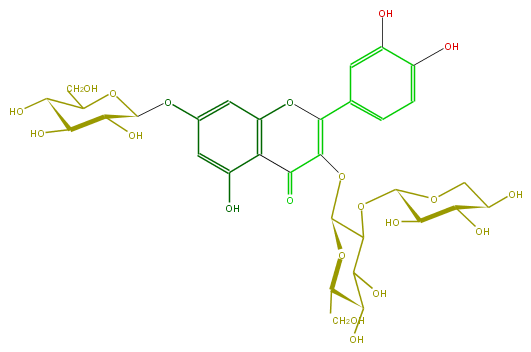
Mol:FL5FACGL0031
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 53 58 0 0 0 0 0 0 0 0999 V2000 | + | 53 58 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.4591 1.2812 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4591 1.2812 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4591 0.4717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4591 0.4717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7581 0.0669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7581 0.0669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0571 0.4717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0571 0.4717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0571 1.2812 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0571 1.2812 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7581 1.6860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7581 1.6860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6440 0.0669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6440 0.0669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3450 0.4717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3450 0.4717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3450 1.2812 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3450 1.2812 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6440 1.6860 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6440 1.6860 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6440 -0.5642 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6440 -0.5642 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0457 1.6857 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0457 1.6857 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7603 1.2733 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7603 1.2733 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4748 1.6857 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4748 1.6857 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4748 2.5107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4748 2.5107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7603 2.9233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7603 2.9233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0457 2.5107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0457 2.5107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1599 1.6857 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1599 1.6857 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8417 -0.0184 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8417 -0.0184 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7581 -0.7422 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7581 -0.7422 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2736 2.9719 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2736 2.9719 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3339 -1.4268 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3339 -1.4268 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5995 -1.0029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5995 -1.0029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8325 -1.8182 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8325 -1.8182 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5995 -2.6386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5995 -2.6386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3339 -3.0626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3339 -3.0626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1009 -2.2472 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1009 -2.2472 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2686 -0.7026 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2686 -0.7026 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6220 -2.6875 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6220 -2.6875 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0911 -3.7481 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0911 -3.7481 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0804 -0.3223 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0804 -0.3223 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6379 -1.0582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6379 -1.0582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4406 -0.7459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4406 -0.7459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2154 -0.7376 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2154 -0.7376 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6525 -0.1746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6525 -0.1746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9501 -0.5452 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9501 -0.5452 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0796 -1.0605 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0796 -1.0605 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9846 -1.2720 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9846 -1.2720 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.7491 -0.4307 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.7491 -0.4307 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9540 1.7531 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9540 1.7531 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4101 1.0352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4101 1.0352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6270 1.3398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6270 1.3398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8713 1.3480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8713 1.3480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4204 1.8971 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4204 1.8971 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1056 1.5356 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1056 1.5356 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.5709 1.4770 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.5709 1.4770 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0871 0.9768 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0871 0.9768 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9898 0.9343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9898 0.9343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7603 3.7481 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7603 3.7481 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5826 -3.3140 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5826 -3.3140 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3302 -3.1749 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3302 -3.1749 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5461 2.0187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5461 2.0187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.7491 2.3940 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.7491 2.3940 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 8 19 1 0 0 0 0 | + | 8 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 21 15 1 0 0 0 0 | + | 21 15 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 23 19 1 0 0 0 0 | + | 23 19 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 32 37 1 0 0 0 0 | + | 32 37 1 0 0 0 0 |
| − | 33 38 1 0 0 0 0 | + | 33 38 1 0 0 0 0 |
| − | 28 31 1 0 0 0 0 | + | 28 31 1 0 0 0 0 |
| − | 34 39 1 0 0 0 0 | + | 34 39 1 0 0 0 0 |
| − | 40 41 1 1 0 0 0 | + | 40 41 1 1 0 0 0 |
| − | 41 42 1 1 0 0 0 | + | 41 42 1 1 0 0 0 |
| − | 43 42 1 1 0 0 0 | + | 43 42 1 1 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 40 1 0 0 0 0 | + | 45 40 1 0 0 0 0 |
| − | 40 46 1 0 0 0 0 | + | 40 46 1 0 0 0 0 |
| − | 41 47 1 0 0 0 0 | + | 41 47 1 0 0 0 0 |
| − | 42 48 1 0 0 0 0 | + | 42 48 1 0 0 0 0 |
| − | 43 18 1 0 0 0 0 | + | 43 18 1 0 0 0 0 |
| − | 16 49 1 0 0 0 0 | + | 16 49 1 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 25 50 1 0 0 0 0 | + | 25 50 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 45 52 1 0 0 0 0 | + | 45 52 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 50 51 | + | M SAL 1 2 50 51 |
| − | M SBL 1 1 56 | + | M SBL 1 1 56 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 56 0.0169 0.6754 | + | M SBV 1 56 0.0169 0.6754 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 52 53 | + | M SAL 2 2 52 53 |
| − | M SBL 2 1 58 | + | M SBL 2 1 58 |
| − | M SMT 2 ^ CH2OH | + | M SMT 2 ^ CH2OH |
| − | M SBV 2 58 0.4406 -0.4831 | + | M SBV 2 58 0.4406 -0.4831 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FACGL0031 | + | ID FL5FACGL0031 |
| − | FORMULA C32H38O21 | + | FORMULA C32H38O21 |
| − | EXACTMASS 758.190558278 | + | EXACTMASS 758.190558278 |
| − | AVERAGEMASS 758.6315199999999 | + | AVERAGEMASS 758.6315199999999 |
| − | SMILES C(CO)(C(O)6)OC(C(C(O)6)OC(O5)C(C(O)C(O)C5)O)OC(C2=O)=C(Oc(c3)c2c(O)cc(OC(C(O)4)OC(CO)C(O)C4O)3)c(c1)ccc(c(O)1)O | + | SMILES C(CO)(C(O)6)OC(C(C(O)6)OC(O5)C(C(O)C(O)C5)O)OC(C2=O)=C(Oc(c3)c2c(O)cc(OC(C(O)4)OC(CO)C(O)C4O)3)c(c1)ccc(c(O)1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
53 58 0 0 0 0 0 0 0 0999 V2000
-1.4591 1.2812 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4591 0.4717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7581 0.0669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0571 0.4717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0571 1.2812 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7581 1.6860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6440 0.0669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3450 0.4717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3450 1.2812 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6440 1.6860 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6440 -0.5642 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0457 1.6857 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7603 1.2733 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4748 1.6857 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4748 2.5107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7603 2.9233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0457 2.5107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1599 1.6857 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8417 -0.0184 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7581 -0.7422 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2736 2.9719 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3339 -1.4268 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5995 -1.0029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8325 -1.8182 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5995 -2.6386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3339 -3.0626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1009 -2.2472 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2686 -0.7026 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6220 -2.6875 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0911 -3.7481 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0804 -0.3223 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6379 -1.0582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4406 -0.7459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.2154 -0.7376 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6525 -0.1746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9501 -0.5452 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0796 -1.0605 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9846 -1.2720 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.7491 -0.4307 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9540 1.7531 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4101 1.0352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6270 1.3398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8713 1.3480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4204 1.8971 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1056 1.5356 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.5709 1.4770 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.0871 0.9768 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9898 0.9343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7603 3.7481 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5826 -3.3140 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3302 -3.1749 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5461 2.0187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.7491 2.3940 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
8 19 1 0 0 0 0
3 20 1 0 0 0 0
21 15 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
23 19 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
32 37 1 0 0 0 0
33 38 1 0 0 0 0
28 31 1 0 0 0 0
34 39 1 0 0 0 0
40 41 1 1 0 0 0
41 42 1 1 0 0 0
43 42 1 1 0 0 0
43 44 1 0 0 0 0
44 45 1 0 0 0 0
45 40 1 0 0 0 0
40 46 1 0 0 0 0
41 47 1 0 0 0 0
42 48 1 0 0 0 0
43 18 1 0 0 0 0
16 49 1 0 0 0 0
50 51 1 0 0 0 0
25 50 1 0 0 0 0
52 53 1 0 0 0 0
45 52 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 50 51
M SBL 1 1 56
M SMT 1 ^ CH2OH
M SBV 1 56 0.0169 0.6754
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 52 53
M SBL 2 1 58
M SMT 2 ^ CH2OH
M SBV 2 58 0.4406 -0.4831
S SKP 5
ID FL5FACGL0031
FORMULA C32H38O21
EXACTMASS 758.190558278
AVERAGEMASS 758.6315199999999
SMILES C(CO)(C(O)6)OC(C(C(O)6)OC(O5)C(C(O)C(O)C5)O)OC(C2=O)=C(Oc(c3)c2c(O)cc(OC(C(O)4)OC(CO)C(O)C4O)3)c(c1)ccc(c(O)1)O
M END