Mol:FL5FAAGS0129
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 61 66 0 0 0 0 0 0 0 0999 V2000 | + | 61 66 0 0 0 0 0 0 0 0999 V2000 |
| − | 2.2416 3.2507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2416 3.2507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2416 2.4258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2416 2.4258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9561 2.0133 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9561 2.0133 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6705 2.4258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6705 2.4258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6705 3.2507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6705 3.2507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9561 3.6632 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9561 3.6632 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5272 2.0133 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5272 2.0133 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8127 2.4258 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8127 2.4258 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0982 2.0133 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0982 2.0133 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0982 1.1883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0982 1.1883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8127 0.7758 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8127 0.7758 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5272 1.1883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5272 1.1883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6162 2.4258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6162 2.4258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3306 2.0133 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3306 2.0133 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3306 1.1883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3306 1.1883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6162 0.7758 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6162 0.7758 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8127 0.0575 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8127 0.0575 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0836 2.3405 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0836 2.3405 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2048 0.7535 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2048 0.7535 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2712 3.5975 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2712 3.5975 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6162 0.0762 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6162 0.0762 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2312 -2.2708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2312 -2.2708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8736 -2.2694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8736 -2.2694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2237 -2.9210 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2237 -2.9210 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2457 -1.6811 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2457 -1.6811 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8547 -1.0992 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8547 -1.0992 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4815 -0.0391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4815 -0.0391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6688 0.1042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6688 0.1042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5923 0.9259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5923 0.9259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1077 1.5940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1077 1.5940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9205 1.4506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9205 1.4506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9971 0.6290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9971 0.6290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7496 0.3734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7496 0.3734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2712 -0.5185 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2712 -0.5185 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4176 -0.8203 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4176 -0.8203 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9242 -0.5655 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9242 -0.5655 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7531 1.3247 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7531 1.3247 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3538 -1.2653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3538 -1.2653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1687 -1.3956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1687 -1.3956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5096 -0.6441 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5096 -0.6441 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1860 -0.1712 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1860 -0.1712 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3711 -0.0408 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3711 -0.0408 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0300 -0.7924 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0300 -0.7924 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0543 -1.8020 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0543 -1.8020 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6242 -1.7469 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6242 -1.7469 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3946 -0.6045 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3946 -0.6045 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0541 -2.1115 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0541 -2.1115 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3487 -2.8824 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3487 -2.8824 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6730 -2.1234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6730 -2.1234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3386 -1.6358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3386 -1.6358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0442 -0.8648 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0442 -0.8648 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7198 -1.6237 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7198 -1.6237 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9449 -2.0143 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9449 -2.0143 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8503 -3.0221 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8503 -3.0221 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7671 -3.0954 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7671 -3.0954 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6745 -2.6309 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6745 -2.6309 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1890 -0.8350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1890 -0.8350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3286 -1.0328 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3286 -1.0328 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8155 -2.9497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8155 -2.9497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9917 -2.9497 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9917 -2.9497 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2274 -3.6632 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2274 -3.6632 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 2 7 1 0 0 0 0 | + | 2 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 11 1 0 0 0 0 | + | 10 11 1 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 7 2 0 0 0 0 | + | 12 7 2 0 0 0 0 |
| − | 9 13 1 0 0 0 0 | + | 9 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 10 1 0 0 0 0 | + | 16 10 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 11 17 2 0 0 0 0 | + | 11 17 2 0 0 0 0 |
| − | 12 19 1 0 0 0 0 | + | 12 19 1 0 0 0 0 |
| − | 5 20 1 0 0 0 0 | + | 5 20 1 0 0 0 0 |
| − | 16 21 1 0 0 0 0 | + | 16 21 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 2 0 0 0 0 | + | 23 24 2 0 0 0 0 |
| − | 23 25 1 0 0 0 0 | + | 23 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 27 28 1 1 0 0 0 | + | 27 28 1 1 0 0 0 |
| − | 28 29 1 1 0 0 0 | + | 28 29 1 1 0 0 0 |
| − | 30 29 1 1 0 0 0 | + | 30 29 1 1 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 27 1 0 0 0 0 | + | 32 27 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 27 35 1 0 0 0 0 | + | 27 35 1 0 0 0 0 |
| − | 28 36 1 0 0 0 0 | + | 28 36 1 0 0 0 0 |
| − | 29 37 1 0 0 0 0 | + | 29 37 1 0 0 0 0 |
| − | 34 26 1 0 0 0 0 | + | 34 26 1 0 0 0 0 |
| − | 18 30 1 0 0 0 0 | + | 18 30 1 0 0 0 0 |
| − | 38 39 1 1 0 0 0 | + | 38 39 1 1 0 0 0 |
| − | 39 40 1 1 0 0 0 | + | 39 40 1 1 0 0 0 |
| − | 41 40 1 1 0 0 0 | + | 41 40 1 1 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 43 38 1 0 0 0 0 | + | 43 38 1 0 0 0 0 |
| − | 38 44 1 0 0 0 0 | + | 38 44 1 0 0 0 0 |
| − | 39 45 1 0 0 0 0 | + | 39 45 1 0 0 0 0 |
| − | 40 46 1 0 0 0 0 | + | 40 46 1 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 48 49 1 1 0 0 0 | + | 48 49 1 1 0 0 0 |
| − | 50 49 1 1 0 0 0 | + | 50 49 1 1 0 0 0 |
| − | 51 50 1 1 0 0 0 | + | 51 50 1 1 0 0 0 |
| − | 51 52 1 0 0 0 0 | + | 51 52 1 0 0 0 0 |
| − | 52 47 1 0 0 0 0 | + | 52 47 1 0 0 0 0 |
| − | 50 53 1 0 0 0 0 | + | 50 53 1 0 0 0 0 |
| − | 49 54 1 0 0 0 0 | + | 49 54 1 0 0 0 0 |
| − | 48 55 1 0 0 0 0 | + | 48 55 1 0 0 0 0 |
| − | 47 56 1 0 0 0 0 | + | 47 56 1 0 0 0 0 |
| − | 51 46 1 0 0 0 0 | + | 51 46 1 0 0 0 0 |
| − | 41 19 1 0 0 0 0 | + | 41 19 1 0 0 0 0 |
| − | 57 58 1 0 0 0 0 | + | 57 58 1 0 0 0 0 |
| − | 43 57 1 0 0 0 0 | + | 43 57 1 0 0 0 0 |
| − | 60 59 2 0 0 0 0 | + | 60 59 2 0 0 0 0 |
| − | 59 61 1 0 0 0 0 | + | 59 61 1 0 0 0 0 |
| − | 22 59 1 0 0 0 0 | + | 22 59 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 57 58 | + | M SAL 1 2 57 58 |
| − | M SBL 1 1 63 | + | M SBL 1 1 63 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 63 0.8410 0.0426 | + | M SBV 1 63 0.8410 0.0426 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 3 59 60 61 | + | M SAL 2 3 59 60 61 |
| − | M SBL 2 1 66 | + | M SBL 2 1 66 |
| − | M SMT 2 COOH | + | M SMT 2 COOH |
| − | M SBV 2 66 -0.4156 0.6788 | + | M SBV 2 66 -0.4156 0.6788 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FAAGS0129 | + | ID FL5FAAGS0129 |
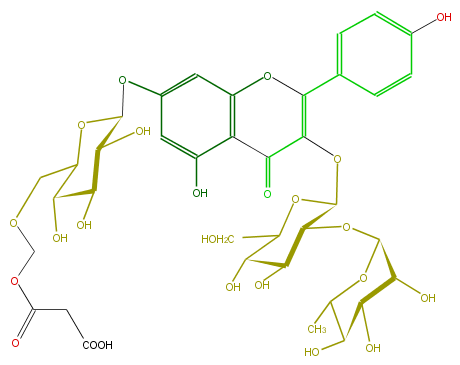
| − | FORMULA C37H44O24 | + | FORMULA C37H44O24 |
| − | EXACTMASS 872.222252336 | + | EXACTMASS 872.222252336 |
| − | AVERAGEMASS 872.7308599999999 | + | AVERAGEMASS 872.7308599999999 |
| − | SMILES O(C(O6)C(O)C(O)C(O)C6C)C(C(O)1)C(OC(=C4c(c5)ccc(c5)O)C(=O)c(c(O4)2)c(cc(OC(O3)C(O)C(C(C3COCOC(=O)CC(O)=O)O)O)c2)O)OC(CO)C1O | + | SMILES O(C(O6)C(O)C(O)C(O)C6C)C(C(O)1)C(OC(=C4c(c5)ccc(c5)O)C(=O)c(c(O4)2)c(cc(OC(O3)C(O)C(C(C3COCOC(=O)CC(O)=O)O)O)c2)O)OC(CO)C1O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
61 66 0 0 0 0 0 0 0 0999 V2000
2.2416 3.2507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2416 2.4258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9561 2.0133 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6705 2.4258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6705 3.2507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9561 3.6632 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5272 2.0133 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8127 2.4258 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0982 2.0133 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0982 1.1883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8127 0.7758 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5272 1.1883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6162 2.4258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3306 2.0133 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3306 1.1883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6162 0.7758 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8127 0.0575 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0836 2.3405 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2048 0.7535 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2712 3.5975 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6162 0.0762 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2312 -2.2708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8736 -2.2694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2237 -2.9210 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2457 -1.6811 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8547 -1.0992 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4815 -0.0391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6688 0.1042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5923 0.9259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1077 1.5940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9205 1.4506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9971 0.6290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7496 0.3734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2712 -0.5185 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4176 -0.8203 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9242 -0.5655 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7531 1.3247 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3538 -1.2653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1687 -1.3956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5096 -0.6441 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1860 -0.1712 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3711 -0.0408 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0300 -0.7924 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0543 -1.8020 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6242 -1.7469 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3946 -0.6045 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0541 -2.1115 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3487 -2.8824 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6730 -2.1234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3386 -1.6358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0442 -0.8648 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7198 -1.6237 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9449 -2.0143 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8503 -3.0221 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7671 -3.0954 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6745 -2.6309 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1890 -0.8350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3286 -1.0328 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8155 -2.9497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9917 -2.9497 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2274 -3.6632 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
2 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 11 1 0 0 0 0
11 12 1 0 0 0 0
12 7 2 0 0 0 0
9 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 10 1 0 0 0 0
14 18 1 0 0 0 0
11 17 2 0 0 0 0
12 19 1 0 0 0 0
5 20 1 0 0 0 0
16 21 1 0 0 0 0
22 23 1 0 0 0 0
23 24 2 0 0 0 0
23 25 1 0 0 0 0
25 26 1 0 0 0 0
27 28 1 1 0 0 0
28 29 1 1 0 0 0
30 29 1 1 0 0 0
30 31 1 0 0 0 0
31 32 1 0 0 0 0
32 27 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
27 35 1 0 0 0 0
28 36 1 0 0 0 0
29 37 1 0 0 0 0
34 26 1 0 0 0 0
18 30 1 0 0 0 0
38 39 1 1 0 0 0
39 40 1 1 0 0 0
41 40 1 1 0 0 0
41 42 1 0 0 0 0
42 43 1 0 0 0 0
43 38 1 0 0 0 0
38 44 1 0 0 0 0
39 45 1 0 0 0 0
40 46 1 0 0 0 0
47 48 1 0 0 0 0
48 49 1 1 0 0 0
50 49 1 1 0 0 0
51 50 1 1 0 0 0
51 52 1 0 0 0 0
52 47 1 0 0 0 0
50 53 1 0 0 0 0
49 54 1 0 0 0 0
48 55 1 0 0 0 0
47 56 1 0 0 0 0
51 46 1 0 0 0 0
41 19 1 0 0 0 0
57 58 1 0 0 0 0
43 57 1 0 0 0 0
60 59 2 0 0 0 0
59 61 1 0 0 0 0
22 59 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 57 58
M SBL 1 1 63
M SMT 1 ^ CH2OH
M SBV 1 63 0.8410 0.0426
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 3 59 60 61
M SBL 2 1 66
M SMT 2 COOH
M SBV 2 66 -0.4156 0.6788
S SKP 5
ID FL5FAAGS0129
FORMULA C37H44O24
EXACTMASS 872.222252336
AVERAGEMASS 872.7308599999999
SMILES O(C(O6)C(O)C(O)C(O)C6C)C(C(O)1)C(OC(=C4c(c5)ccc(c5)O)C(=O)c(c(O4)2)c(cc(OC(O3)C(O)C(C(C3COCOC(=O)CC(O)=O)O)O)c2)O)OC(CO)C1O
M END