Mol:FL5FAAGS0074
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | 1.9122 2.3818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9122 2.3818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9122 1.5568 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9122 1.5568 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6267 1.1443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6267 1.1443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3411 1.5568 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3411 1.5568 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3411 2.3818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3411 2.3818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6267 2.7943 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6267 2.7943 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1977 1.1443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1977 1.1443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4832 1.5568 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4832 1.5568 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2312 1.1443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2312 1.1443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2312 0.3193 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2312 0.3193 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4832 -0.0932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4832 -0.0932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1977 0.3193 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1977 0.3193 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9457 1.5568 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9457 1.5568 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6602 1.1443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6602 1.1443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6602 0.3193 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6602 0.3193 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9457 -0.0932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9457 -0.0932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4832 -0.8114 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4832 -0.8114 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3746 1.5568 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3746 1.5568 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8040 -0.0307 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8040 -0.0307 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9419 2.7287 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9419 2.7287 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9457 -0.7927 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9457 -0.7927 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4588 -0.5638 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4588 -0.5638 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0463 -1.2783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0463 -1.2783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2482 -1.0692 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2482 -1.0692 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4550 -1.2960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4550 -1.2960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8674 -0.5814 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8674 -0.5814 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6656 -0.7905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6656 -0.7905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8225 -0.2048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8225 -0.2048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3022 0.0721 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3022 0.0721 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9419 -1.0469 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9419 -1.0469 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4948 -1.0193 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4948 -1.0193 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4196 -1.7089 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4196 -1.7089 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0726 -2.1122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0726 -2.1122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7346 -2.2826 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7346 -2.2826 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1123 -1.5491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1123 -1.5491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8110 -1.1104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8110 -1.1104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0038 -0.9399 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0038 -0.9399 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6260 -1.6734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6260 -1.6734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0854 -1.3989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0854 -1.3989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4411 -1.5708 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4411 -1.5708 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0359 -2.7943 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0359 -2.7943 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2423 -2.4433 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2423 -2.4433 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4737 -2.1041 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4737 -2.1041 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 2 7 1 0 0 0 0 | + | 2 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 11 1 0 0 0 0 | + | 10 11 1 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 7 2 0 0 0 0 | + | 12 7 2 0 0 0 0 |
| − | 9 13 1 0 0 0 0 | + | 9 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 10 1 0 0 0 0 | + | 16 10 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 11 17 2 0 0 0 0 | + | 11 17 2 0 0 0 0 |
| − | 12 19 1 0 0 0 0 | + | 12 19 1 0 0 0 0 |
| − | 5 20 1 0 0 0 0 | + | 5 20 1 0 0 0 0 |
| − | 16 21 1 0 0 0 0 | + | 16 21 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 22 30 1 0 0 0 0 | + | 22 30 1 0 0 0 0 |
| − | 23 31 1 0 0 0 0 | + | 23 31 1 0 0 0 0 |
| − | 24 32 1 0 0 0 0 | + | 24 32 1 0 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 36 35 1 1 0 0 0 | + | 36 35 1 1 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 33 1 0 0 0 0 | + | 38 33 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 33 41 1 0 0 0 0 | + | 33 41 1 0 0 0 0 |
| − | 34 42 1 0 0 0 0 | + | 34 42 1 0 0 0 0 |
| − | 35 43 1 0 0 0 0 | + | 35 43 1 0 0 0 0 |
| − | 36 19 1 0 0 0 0 | + | 36 19 1 0 0 0 0 |
| − | 25 21 1 0 0 0 0 | + | 25 21 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FAAGS0074 | + | ID FL5FAAGS0074 |
| − | KNApSAcK_ID C00013741 | + | KNApSAcK_ID C00013741 |
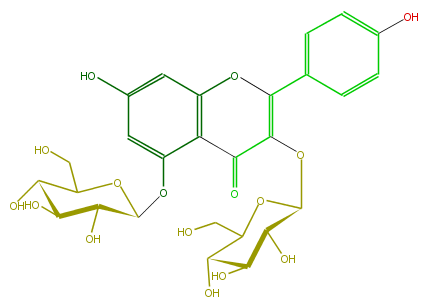
| − | NAME Kaempferol 3,5-diglucoside;3,5-Bis(beta-D-glucopyranosyloxy)-7-hydroxy-2-(4-hydroxyphenyl)-4H-1-benzopyran-4-one | + | NAME Kaempferol 3,5-diglucoside;3,5-Bis(beta-D-glucopyranosyloxy)-7-hydroxy-2-(4-hydroxyphenyl)-4H-1-benzopyran-4-one |
| − | CAS_RN 205103-97-5 | + | CAS_RN 205103-97-5 |
| − | FORMULA C27H30O16 | + | FORMULA C27H30O16 |
| − | EXACTMASS 610.153384912 | + | EXACTMASS 610.153384912 |
| − | AVERAGEMASS 610.5175 | + | AVERAGEMASS 610.5175 |
| − | SMILES O(C=1c(c5)ccc(c5)O)c(c4)c(c(cc(O)4)OC(O3)C(C(O)C(C3CO)O)O)C(=O)C1OC(C(O)2)OC(C(C(O)2)O)CO | + | SMILES O(C=1c(c5)ccc(c5)O)c(c4)c(c(cc(O)4)OC(O3)C(C(O)C(C3CO)O)O)C(=O)C1OC(C(O)2)OC(C(C(O)2)O)CO |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
1.9122 2.3818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9122 1.5568 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6267 1.1443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3411 1.5568 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3411 2.3818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6267 2.7943 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1977 1.1443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4832 1.5568 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2312 1.1443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2312 0.3193 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4832 -0.0932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1977 0.3193 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9457 1.5568 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6602 1.1443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6602 0.3193 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9457 -0.0932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4832 -0.8114 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3746 1.5568 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8040 -0.0307 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9419 2.7287 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9457 -0.7927 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4588 -0.5638 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0463 -1.2783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2482 -1.0692 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4550 -1.2960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8674 -0.5814 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6656 -0.7905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8225 -0.2048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3022 0.0721 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9419 -1.0469 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4948 -1.0193 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4196 -1.7089 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0726 -2.1122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7346 -2.2826 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1123 -1.5491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8110 -1.1104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0038 -0.9399 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6260 -1.6734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0854 -1.3989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4411 -1.5708 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0359 -2.7943 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2423 -2.4433 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4737 -2.1041 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
2 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 11 1 0 0 0 0
11 12 1 0 0 0 0
12 7 2 0 0 0 0
9 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 10 1 0 0 0 0
14 18 1 0 0 0 0
11 17 2 0 0 0 0
12 19 1 0 0 0 0
5 20 1 0 0 0 0
16 21 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
27 28 1 0 0 0 0
28 29 1 0 0 0 0
22 30 1 0 0 0 0
23 31 1 0 0 0 0
24 32 1 0 0 0 0
33 34 1 1 0 0 0
34 35 1 1 0 0 0
36 35 1 1 0 0 0
36 37 1 0 0 0 0
37 38 1 0 0 0 0
38 33 1 0 0 0 0
38 39 1 0 0 0 0
39 40 1 0 0 0 0
33 41 1 0 0 0 0
34 42 1 0 0 0 0
35 43 1 0 0 0 0
36 19 1 0 0 0 0
25 21 1 0 0 0 0
S SKP 8
ID FL5FAAGS0074
KNApSAcK_ID C00013741
NAME Kaempferol 3,5-diglucoside;3,5-Bis(beta-D-glucopyranosyloxy)-7-hydroxy-2-(4-hydroxyphenyl)-4H-1-benzopyran-4-one
CAS_RN 205103-97-5
FORMULA C27H30O16
EXACTMASS 610.153384912
AVERAGEMASS 610.5175
SMILES O(C=1c(c5)ccc(c5)O)c(c4)c(c(cc(O)4)OC(O3)C(C(O)C(C3CO)O)O)C(=O)C1OC(C(O)2)OC(C(C(O)2)O)CO
M END