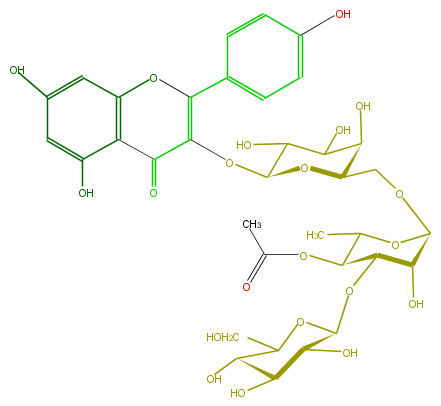
Mol:FL5FAAGA0039
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 56 61 0 0 0 0 0 0 0 0999 V2000 | + | 56 61 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.5019 2.0971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5019 2.0971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5019 1.2721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5019 1.2721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7874 0.8595 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7874 0.8595 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0729 1.2721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0729 1.2721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0729 2.0971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0729 2.0971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7874 2.5097 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7874 2.5097 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3583 0.8595 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3583 0.8595 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6438 1.2721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6438 1.2721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6438 2.0971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6438 2.0971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3583 2.5097 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3583 2.5097 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3583 0.2161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3583 0.2161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1116 2.5034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1116 2.5034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8399 2.0830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8399 2.0830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5680 2.5034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5680 2.5034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5680 3.3444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5680 3.3444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8399 3.7649 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8399 3.7649 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1116 3.3444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1116 3.3444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7874 0.2212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7874 0.2212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3476 3.7944 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3476 3.7944 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1642 2.6800 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1642 2.6800 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1646 0.8053 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1646 0.8053 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3005 1.1789 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3005 1.1789 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9141 0.5097 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9141 0.5097 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6572 0.7220 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6572 0.7220 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4047 0.5097 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4047 0.5097 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7912 1.1789 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7912 1.1789 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0481 0.9666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0481 0.9666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5188 1.1669 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5188 1.1669 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3502 1.4717 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3502 1.4717 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7513 1.9460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7513 1.9460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0747 0.6720 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0747 0.6720 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5478 0.1971 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5478 0.1971 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7695 -0.6199 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7695 -0.6199 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4080 -1.2461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4080 -1.2461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1031 -1.0474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1031 -1.0474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8026 -1.2461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8026 -1.2461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1642 -0.6199 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1642 -0.6199 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4689 -0.8185 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4689 -0.8185 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5579 -1.7425 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5579 -1.7425 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6335 -1.0369 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6335 -1.0369 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9862 -0.6271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9862 -0.6271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8211 -2.0077 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8211 -2.0077 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2923 -3.1070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2923 -3.1070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0602 -3.5127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0602 -3.5127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6205 -2.9275 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6205 -2.9275 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2775 -2.6122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2775 -2.6122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5739 -2.3559 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5739 -2.3559 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1223 -2.9513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1223 -2.9513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2319 -3.5096 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2319 -3.5096 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4016 -3.7944 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4016 -3.7944 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4859 -2.9918 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4859 -2.9918 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8498 -1.0369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8498 -1.0369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4865 -1.6661 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4865 -1.6661 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4888 -0.4117 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4888 -0.4117 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3980 -2.6606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3980 -2.6606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7943 -2.8758 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7943 -2.8758 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 1 20 1 0 0 0 0 | + | 1 20 1 0 0 0 0 |
| − | 21 8 1 0 0 0 0 | + | 21 8 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 25 31 1 0 0 0 0 | + | 25 31 1 0 0 0 0 |
| − | 23 21 1 0 0 0 0 | + | 23 21 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 35 36 1 1 0 0 0 | + | 35 36 1 1 0 0 0 |
| − | 37 36 1 1 0 0 0 | + | 37 36 1 1 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 33 1 0 0 0 0 | + | 38 33 1 0 0 0 0 |
| − | 35 39 1 0 0 0 0 | + | 35 39 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 33 41 1 0 0 0 0 | + | 33 41 1 0 0 0 0 |
| − | 36 42 1 0 0 0 0 | + | 36 42 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 43 44 1 1 0 0 0 | + | 43 44 1 1 0 0 0 |
| − | 44 45 1 1 0 0 0 | + | 44 45 1 1 0 0 0 |
| − | 46 45 1 1 0 0 0 | + | 46 45 1 1 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 48 43 1 0 0 0 0 | + | 48 43 1 0 0 0 0 |
| − | 43 49 1 0 0 0 0 | + | 43 49 1 0 0 0 0 |
| − | 44 50 1 0 0 0 0 | + | 44 50 1 0 0 0 0 |
| − | 45 51 1 0 0 0 0 | + | 45 51 1 0 0 0 0 |
| − | 46 39 1 0 0 0 0 | + | 46 39 1 0 0 0 0 |
| − | 40 52 1 0 0 0 0 | + | 40 52 1 0 0 0 0 |
| − | 52 53 2 0 0 0 0 | + | 52 53 2 0 0 0 0 |
| − | 52 54 1 0 0 0 0 | + | 52 54 1 0 0 0 0 |
| − | 55 56 1 0 0 0 0 | + | 55 56 1 0 0 0 0 |
| − | 48 55 1 0 0 0 0 | + | 48 55 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 55 56 | + | M SAL 1 2 55 56 |
| − | M SBL 1 1 61 | + | M SBL 1 1 61 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 61 0.7244 -0.2907 | + | M SBV 1 61 0.7244 -0.2907 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FAAGA0039 | + | ID FL5FAAGA0039 |
| − | FORMULA C35H42O21 | + | FORMULA C35H42O21 |
| − | EXACTMASS 798.2218584059999 | + | EXACTMASS 798.2218584059999 |
| − | AVERAGEMASS 798.69538 | + | AVERAGEMASS 798.69538 |
| − | SMILES c(O)(c1)ccc(C(=C(OC(O4)C(O)C(C(O)C4COC(O6)C(O)C(C(C6C)OC(C)=O)OC(O5)C(C(C(C5CO)O)O)O)O)2)Oc(c3)c(c(cc3O)O)C2=O)c1 | + | SMILES c(O)(c1)ccc(C(=C(OC(O4)C(O)C(C(O)C4COC(O6)C(O)C(C(C6C)OC(C)=O)OC(O5)C(C(C(C5CO)O)O)O)O)2)Oc(c3)c(c(cc3O)O)C2=O)c1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
56 61 0 0 0 0 0 0 0 0999 V2000
-3.5019 2.0971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5019 1.2721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7874 0.8595 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0729 1.2721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0729 2.0971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7874 2.5097 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3583 0.8595 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6438 1.2721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6438 2.0971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3583 2.5097 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3583 0.2161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1116 2.5034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8399 2.0830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5680 2.5034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5680 3.3444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8399 3.7649 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1116 3.3444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7874 0.2212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3476 3.7944 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1642 2.6800 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1646 0.8053 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3005 1.1789 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9141 0.5097 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6572 0.7220 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4047 0.5097 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7912 1.1789 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0481 0.9666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5188 1.1669 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3502 1.4717 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7513 1.9460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0747 0.6720 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5478 0.1971 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7695 -0.6199 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4080 -1.2461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1031 -1.0474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8026 -1.2461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1642 -0.6199 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4689 -0.8185 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5579 -1.7425 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6335 -1.0369 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9862 -0.6271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8211 -2.0077 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2923 -3.1070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0602 -3.5127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6205 -2.9275 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2775 -2.6122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5739 -2.3559 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1223 -2.9513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2319 -3.5096 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4016 -3.7944 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4859 -2.9918 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8498 -1.0369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4865 -1.6661 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4888 -0.4117 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3980 -2.6606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7943 -2.8758 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
4 3 1 0 0 0 0
1 20 1 0 0 0 0
21 8 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
25 31 1 0 0 0 0
23 21 1 0 0 0 0
31 32 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 1 0 0 0
35 36 1 1 0 0 0
37 36 1 1 0 0 0
37 38 1 0 0 0 0
38 33 1 0 0 0 0
35 39 1 0 0 0 0
34 40 1 0 0 0 0
33 41 1 0 0 0 0
36 42 1 0 0 0 0
37 32 1 0 0 0 0
43 44 1 1 0 0 0
44 45 1 1 0 0 0
46 45 1 1 0 0 0
46 47 1 0 0 0 0
47 48 1 0 0 0 0
48 43 1 0 0 0 0
43 49 1 0 0 0 0
44 50 1 0 0 0 0
45 51 1 0 0 0 0
46 39 1 0 0 0 0
40 52 1 0 0 0 0
52 53 2 0 0 0 0
52 54 1 0 0 0 0
55 56 1 0 0 0 0
48 55 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 55 56
M SBL 1 1 61
M SMT 1 ^ CH2OH
M SBV 1 61 0.7244 -0.2907
S SKP 5
ID FL5FAAGA0039
FORMULA C35H42O21
EXACTMASS 798.2218584059999
AVERAGEMASS 798.69538
SMILES c(O)(c1)ccc(C(=C(OC(O4)C(O)C(C(O)C4COC(O6)C(O)C(C(C6C)OC(C)=O)OC(O5)C(C(C(C5CO)O)O)O)O)2)Oc(c3)c(c(cc3O)O)C2=O)c1
M END