Mol:FL3FGCGS0006
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 37 40 0 0 0 0 0 0 0 0999 V2000 | + | 37 40 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.2974 -0.3905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2974 -0.3905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2974 -0.9525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2974 -0.9525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8463 -1.2130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8463 -1.2130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3953 -0.9525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3953 -0.9525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3953 -0.4317 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3953 -0.4317 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8463 -0.1713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8463 -0.1713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9442 -1.2130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9442 -1.2130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4931 -0.9525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4931 -0.9525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4931 -0.4317 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4931 -0.4317 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9442 -0.1713 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9442 -0.1713 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7725 -1.5809 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7725 -1.5809 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0422 -0.1714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0422 -0.1714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5825 -0.4368 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5825 -0.4368 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1228 -0.1714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1228 -0.1714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1228 0.3595 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1228 0.3595 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5825 0.6249 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5825 0.6249 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0422 0.3595 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0422 0.3595 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8463 -1.7327 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8463 -1.7327 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7502 -0.0916 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7502 -0.0916 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8772 0.3595 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8772 0.3595 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6774 1.4141 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.6774 1.4141 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.2898 0.7426 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.2898 0.7426 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.0352 0.9557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0352 0.9557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7852 0.7426 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.7852 0.7426 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.1730 1.4141 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.1730 1.4141 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.4274 1.2011 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.4274 1.2011 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.9575 1.2211 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9575 1.2211 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7344 1.7327 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7344 1.7327 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7323 1.0911 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7323 1.0911 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0119 -0.8205 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0119 -0.8205 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9953 -0.6388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9953 -0.6388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4143 1.2060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4143 1.2060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1363 2.1666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1363 2.1666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6075 0.3791 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6075 0.3791 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2095 1.2965 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2095 1.2965 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2974 0.7693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2974 0.7693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0119 0.3568 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0119 0.3568 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 20 15 1 0 0 0 0 | + | 20 15 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 26 28 1 0 0 0 0 | + | 26 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 22 20 1 0 0 0 0 | + | 22 20 1 0 0 0 0 |
| − | 2 30 1 0 0 0 0 | + | 2 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 16 32 1 0 0 0 0 | + | 16 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 6 34 1 0 0 0 0 | + | 6 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 24 36 1 0 0 0 0 | + | 24 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 36 37 | + | M SAL 4 2 36 37 |
| − | M SBL 4 1 39 | + | M SBL 4 1 39 |
| − | M SMT 4 CH2OH | + | M SMT 4 CH2OH |
| − | M SVB 4 39 3.2974 0.7693 | + | M SVB 4 39 3.2974 0.7693 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 34 35 | + | M SAL 3 2 34 35 |
| − | M SBL 3 1 37 | + | M SBL 3 1 37 |
| − | M SMT 3 OCH3 | + | M SMT 3 OCH3 |
| − | M SVB 3 37 -2.6075 0.3791 | + | M SVB 3 37 -2.6075 0.3791 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 32 33 | + | M SAL 2 2 32 33 |
| − | M SBL 2 1 35 | + | M SBL 2 1 35 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 35 -0.4143 1.206 | + | M SVB 2 35 -0.4143 1.206 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 30 31 | + | M SAL 1 2 30 31 |
| − | M SBL 1 1 33 | + | M SBL 1 1 33 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 33 -4.0119 -0.8205 | + | M SVB 1 33 -4.0119 -0.8205 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FGCGS0006 | + | ID FL3FGCGS0006 |
| − | KNApSAcK_ID C00004442 | + | KNApSAcK_ID C00004442 |
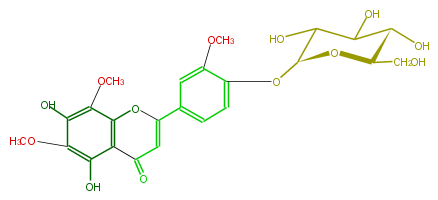
| − | NAME 5,7,4'-Trihydroxy-6,8,3'-trimethoxyflavone 4'-glucoside | + | NAME 5,7,4'-Trihydroxy-6,8,3'-trimethoxyflavone 4'-glucoside |
| − | CAS_RN 70575-17-6 | + | CAS_RN 70575-17-6 |
| − | FORMULA C24H26O13 | + | FORMULA C24H26O13 |
| − | EXACTMASS 522.137340918 | + | EXACTMASS 522.137340918 |
| − | AVERAGEMASS 522.45544 | + | AVERAGEMASS 522.45544 |
| − | SMILES C(=O)(C=2)c(c1O)c(OC2c(c4)ccc(c(OC)4)O[C@@H](C3O)O[C@@H]([C@@H](C(O)3)O)CO)c(OC)c(O)c(OC)1 | + | SMILES C(=O)(C=2)c(c1O)c(OC2c(c4)ccc(c(OC)4)O[C@@H](C3O)O[C@@H]([C@@H](C(O)3)O)CO)c(OC)c(O)c(OC)1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
37 40 0 0 0 0 0 0 0 0999 V2000
-3.2974 -0.3905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2974 -0.9525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8463 -1.2130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3953 -0.9525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3953 -0.4317 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8463 -0.1713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9442 -1.2130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4931 -0.9525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4931 -0.4317 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9442 -0.1713 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7725 -1.5809 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0422 -0.1714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5825 -0.4368 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1228 -0.1714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1228 0.3595 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5825 0.6249 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0422 0.3595 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8463 -1.7327 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7502 -0.0916 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8772 0.3595 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6774 1.4141 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.2898 0.7426 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.0352 0.9557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7852 0.7426 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.1730 1.4141 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.4274 1.2011 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.9575 1.2211 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7344 1.7327 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7323 1.0911 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0119 -0.8205 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9953 -0.6388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4143 1.2060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1363 2.1666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6075 0.3791 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2095 1.2965 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2974 0.7693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0119 0.3568 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
20 15 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
26 28 1 0 0 0 0
25 29 1 0 0 0 0
22 20 1 0 0 0 0
2 30 1 0 0 0 0
30 31 1 0 0 0 0
16 32 1 0 0 0 0
32 33 1 0 0 0 0
6 34 1 0 0 0 0
34 35 1 0 0 0 0
24 36 1 0 0 0 0
36 37 1 0 0 0 0
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 36 37
M SBL 4 1 39
M SMT 4 CH2OH
M SVB 4 39 3.2974 0.7693
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 34 35
M SBL 3 1 37
M SMT 3 OCH3
M SVB 3 37 -2.6075 0.3791
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 32 33
M SBL 2 1 35
M SMT 2 OCH3
M SVB 2 35 -0.4143 1.206
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 30 31
M SBL 1 1 33
M SMT 1 OCH3
M SVB 1 33 -4.0119 -0.8205
S SKP 8
ID FL3FGCGS0006
KNApSAcK_ID C00004442
NAME 5,7,4'-Trihydroxy-6,8,3'-trimethoxyflavone 4'-glucoside
CAS_RN 70575-17-6
FORMULA C24H26O13
EXACTMASS 522.137340918
AVERAGEMASS 522.45544
SMILES C(=O)(C=2)c(c1O)c(OC2c(c4)ccc(c(OC)4)O[C@@H](C3O)O[C@@H]([C@@H](C(O)3)O)CO)c(OC)c(O)c(OC)1
M END