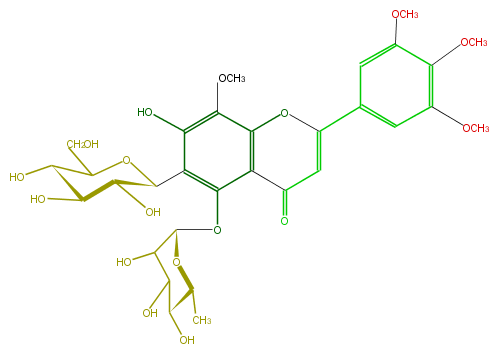
Mol:FL3FFGDS0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 48 52 0 0 0 0 0 0 0 0999 V2000 | + | 48 52 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.8182 0.3691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8182 0.3691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8182 -0.4560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8182 -0.4560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1038 -0.8685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1038 -0.8685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3893 -0.4560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3893 -0.4560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3893 0.3691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3893 0.3691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1038 0.7816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1038 0.7816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3251 -0.8685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3251 -0.8685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0396 -0.4560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0396 -0.4560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0396 0.3691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0396 0.3691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3251 0.7816 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3251 0.7816 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3251 -1.5117 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3251 -1.5117 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1038 -1.6931 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1038 -1.6931 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9128 0.9139 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9128 0.9139 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6657 0.4791 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6657 0.4791 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4185 0.9139 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4185 0.9139 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4185 1.7832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4185 1.7832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6657 2.2178 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6657 2.2178 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9128 1.7832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9128 1.7832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6232 -0.3368 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6232 -0.3368 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9609 -1.0785 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9609 -1.0785 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2987 -0.6812 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2987 -0.6812 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3981 -0.7871 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3981 -0.7871 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0339 -0.2044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0339 -0.2044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7490 -0.5488 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7490 -0.5488 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2696 -0.5763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2696 -0.5763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8211 -1.0374 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8211 -1.0374 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5532 0.7934 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5532 0.7934 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5493 -1.3089 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5493 -1.3089 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1208 -3.4491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1208 -3.4491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6431 -2.9714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6431 -2.9714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9777 -2.3846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9777 -2.3846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9777 -1.7092 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9777 -1.7092 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4555 -2.1869 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4555 -2.1869 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1208 -2.7738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1208 -2.7738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8216 -4.0279 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8216 -4.0279 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5668 -3.5942 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5668 -3.5942 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6075 -3.4612 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6075 -3.4612 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0065 -2.3807 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0065 -2.3807 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1815 0.4560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1815 0.4560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2938 -0.1862 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2938 -0.1862 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1165 1.5161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1165 1.5161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5558 2.6715 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5558 2.6715 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6720 2.8809 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6720 2.8809 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2497 4.0279 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2497 4.0279 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3336 0.1226 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3336 0.1226 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2938 0.5704 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2938 0.5704 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1196 2.2796 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1196 2.2796 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2318 2.9219 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2318 2.9219 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 3 12 1 0 0 0 0 | + | 3 12 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 2 0 0 0 0 | + | 17 18 2 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 9 13 1 0 0 0 0 | + | 9 13 1 0 0 0 0 |
| − | 19 20 1 1 0 0 0 | + | 19 20 1 1 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 19 25 1 0 0 0 0 | + | 19 25 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 2 22 1 0 0 0 0 | + | 2 22 1 0 0 0 0 |
| − | 1 27 1 0 0 0 0 | + | 1 27 1 0 0 0 0 |
| − | 21 28 1 0 0 0 0 | + | 21 28 1 0 0 0 0 |
| − | 29 30 1 1 0 0 0 | + | 29 30 1 1 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 29 1 0 0 0 0 | + | 34 29 1 0 0 0 0 |
| − | 29 35 1 0 0 0 0 | + | 29 35 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 34 37 1 0 0 0 0 | + | 34 37 1 0 0 0 0 |
| − | 32 12 1 0 0 0 0 | + | 32 12 1 0 0 0 0 |
| − | 33 38 1 0 0 0 0 | + | 33 38 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 15 39 1 0 0 0 0 | + | 15 39 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 6 41 1 0 0 0 0 | + | 6 41 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 17 43 1 0 0 0 0 | + | 17 43 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 24 45 1 0 0 0 0 | + | 24 45 1 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 16 47 1 0 0 0 0 | + | 16 47 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 39 40 | + | M SAL 1 2 39 40 |
| − | M SBL 1 1 44 | + | M SBL 1 1 44 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 44 -0.7630 0.4579 | + | M SBV 1 44 -0.7630 0.4579 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 41 42 | + | M SAL 2 2 41 42 |
| − | M SBL 2 1 46 | + | M SBL 2 1 46 |
| − | M SMT 2 ^ OCH3 | + | M SMT 2 ^ OCH3 |
| − | M SBV 2 46 0.0127 -0.7345 | + | M SBV 2 46 0.0127 -0.7345 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 43 44 | + | M SAL 3 2 43 44 |
| − | M SBL 3 1 48 | + | M SBL 3 1 48 |
| − | M SMT 3 OCH3 | + | M SMT 3 OCH3 |
| − | M SBV 3 48 -0.0063 -0.6631 | + | M SBV 3 48 -0.0063 -0.6631 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 45 46 | + | M SAL 4 2 45 46 |
| − | M SBL 4 1 50 | + | M SBL 4 1 50 |
| − | M SMT 4 ^ CH2OH | + | M SMT 4 ^ CH2OH |
| − | M SBV 4 50 0.5846 -0.6714 | + | M SBV 4 50 0.5846 -0.6714 |
| − | M STY 1 5 SUP | + | M STY 1 5 SUP |
| − | M SLB 1 5 5 | + | M SLB 1 5 5 |
| − | M SAL 5 2 47 48 | + | M SAL 5 2 47 48 |
| − | M SBL 5 1 52 | + | M SBL 5 1 52 |
| − | M SMT 5 OCH3 | + | M SMT 5 OCH3 |
| − | M SBV 5 52 -0.7011 -0.4964 | + | M SBV 5 52 -0.7011 -0.4964 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FFGDS0001 | + | ID FL3FFGDS0001 |
| − | FORMULA C31H38O17 | + | FORMULA C31H38O17 |
| − | EXACTMASS 682.21089979 | + | EXACTMASS 682.21089979 |
| − | AVERAGEMASS 682.62322 | + | AVERAGEMASS 682.62322 |
| − | SMILES OC(C1O)C(OC(c(c5O)c(c(c4c(OC)5)C(=O)C=C(O4)c(c3)cc(c(c(OC)3)OC)OC)OC(O2)C(C(O)C(C(C)2)O)O)C1O)CO | + | SMILES OC(C1O)C(OC(c(c5O)c(c(c4c(OC)5)C(=O)C=C(O4)c(c3)cc(c(c(OC)3)OC)OC)OC(O2)C(C(O)C(C(C)2)O)O)C1O)CO |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
48 52 0 0 0 0 0 0 0 0999 V2000
-1.8182 0.3691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8182 -0.4560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1038 -0.8685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3893 -0.4560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3893 0.3691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1038 0.7816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3251 -0.8685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0396 -0.4560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0396 0.3691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3251 0.7816 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3251 -1.5117 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1038 -1.6931 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9128 0.9139 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6657 0.4791 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4185 0.9139 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4185 1.7832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6657 2.2178 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9128 1.7832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6232 -0.3368 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9609 -1.0785 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2987 -0.6812 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3981 -0.7871 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0339 -0.2044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7490 -0.5488 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.2696 -0.5763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8211 -1.0374 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5532 0.7934 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5493 -1.3089 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1208 -3.4491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6431 -2.9714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9777 -2.3846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9777 -1.7092 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4555 -2.1869 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1208 -2.7738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8216 -4.0279 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5668 -3.5942 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6075 -3.4612 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0065 -2.3807 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1815 0.4560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.2938 -0.1862 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1165 1.5161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5558 2.6715 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6720 2.8809 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2497 4.0279 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3336 0.1226 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.2938 0.5704 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1196 2.2796 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.2318 2.9219 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
3 12 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 17 1 0 0 0 0
17 18 2 0 0 0 0
18 13 1 0 0 0 0
9 13 1 0 0 0 0
19 20 1 1 0 0 0
20 21 1 1 0 0 0
22 21 1 1 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 19 1 0 0 0 0
19 25 1 0 0 0 0
20 26 1 0 0 0 0
2 22 1 0 0 0 0
1 27 1 0 0 0 0
21 28 1 0 0 0 0
29 30 1 1 0 0 0
30 31 1 1 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 29 1 0 0 0 0
29 35 1 0 0 0 0
30 36 1 0 0 0 0
34 37 1 0 0 0 0
32 12 1 0 0 0 0
33 38 1 0 0 0 0
39 40 1 0 0 0 0
15 39 1 0 0 0 0
41 42 1 0 0 0 0
6 41 1 0 0 0 0
43 44 1 0 0 0 0
17 43 1 0 0 0 0
45 46 1 0 0 0 0
24 45 1 0 0 0 0
47 48 1 0 0 0 0
16 47 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 39 40
M SBL 1 1 44
M SMT 1 OCH3
M SBV 1 44 -0.7630 0.4579
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 41 42
M SBL 2 1 46
M SMT 2 ^ OCH3
M SBV 2 46 0.0127 -0.7345
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 43 44
M SBL 3 1 48
M SMT 3 OCH3
M SBV 3 48 -0.0063 -0.6631
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 45 46
M SBL 4 1 50
M SMT 4 ^ CH2OH
M SBV 4 50 0.5846 -0.6714
M STY 1 5 SUP
M SLB 1 5 5
M SAL 5 2 47 48
M SBL 5 1 52
M SMT 5 OCH3
M SBV 5 52 -0.7011 -0.4964
S SKP 5
ID FL3FFGDS0001
FORMULA C31H38O17
EXACTMASS 682.21089979
AVERAGEMASS 682.62322
SMILES OC(C1O)C(OC(c(c5O)c(c(c4c(OC)5)C(=O)C=C(O4)c(c3)cc(c(c(OC)3)OC)OC)OC(O2)C(C(O)C(C(C)2)O)O)C1O)CO
M END