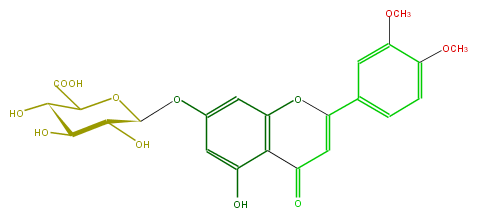
Mol:FL3FAFGS0002
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 35 38 0 0 0 0 0 0 0 0999 V2000 | + | 35 38 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.2980 -0.8973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2980 -0.8973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2980 -1.7067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2980 -1.7067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5969 -2.1116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5969 -2.1116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1042 -1.7067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1042 -1.7067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1042 -0.8973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1042 -0.8973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5969 -0.4925 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5969 -0.4925 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8052 -2.1116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8052 -2.1116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5063 -1.7067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5063 -1.7067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5063 -0.8973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5063 -0.8973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8052 -0.4925 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8052 -0.4925 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8052 -2.9073 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8052 -2.9073 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2071 -0.4927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2071 -0.4927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9216 -0.9052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9216 -0.9052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6361 -0.4927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6361 -0.4927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6361 0.3324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6361 0.3324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9216 0.7449 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9216 0.7449 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2071 0.3324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2071 0.3324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5969 -2.9193 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5969 -2.9193 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9975 -0.4933 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9975 -0.4933 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9868 -0.6057 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9868 -0.6057 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4259 -1.3462 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4259 -1.3462 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6181 -1.0321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6181 -1.0321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8386 -1.0237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8386 -1.0237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4049 -0.4571 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4049 -0.4571 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2302 -0.7534 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2302 -0.7534 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.6333 -0.8249 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.6333 -0.8249 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0515 -1.2664 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0515 -1.2664 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8650 -1.5423 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8650 -1.5423 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9216 1.5095 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9216 1.5095 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5758 2.9193 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5758 2.9193 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8959 -0.1238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8959 -0.1238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.7933 -0.1238 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.7933 -0.1238 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4455 0.6563 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4455 0.6563 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2391 0.6806 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2391 0.6806 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.7933 0.6806 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.7933 0.6806 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 19 1 0 0 0 0 | + | 23 19 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 16 29 1 0 0 0 0 | + | 16 29 1 0 0 0 0 |
| − | 31 32 2 0 0 0 0 | + | 31 32 2 0 0 0 0 |
| − | 31 33 1 0 0 0 0 | + | 31 33 1 0 0 0 0 |
| − | 25 31 1 0 0 0 0 | + | 25 31 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 15 34 1 0 0 0 0 | + | 15 34 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 29 30 | + | M SAL 1 2 29 30 |
| − | M SBL 1 1 33 | + | M SBL 1 1 33 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 33 0.0000 -0.7645 | + | M SBV 1 33 0.0000 -0.7645 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 3 31 32 33 | + | M SAL 2 3 31 32 33 |
| − | M SBL 2 1 36 | + | M SBL 2 1 36 |
| − | M SMT 2 ^ COOH | + | M SMT 2 ^ COOH |
| − | M SBV 2 36 0.6657 -0.6296 | + | M SBV 2 36 0.6657 -0.6296 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 34 35 | + | M SAL 3 2 34 35 |
| − | M SBL 3 1 38 | + | M SBL 3 1 38 |
| − | M SMT 3 OCH3 | + | M SMT 3 OCH3 |
| − | M SBV 3 38 -0.6030 -0.3482 | + | M SBV 3 38 -0.6030 -0.3482 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FAFGS0002 | + | ID FL3FAFGS0002 |
| − | FORMULA C23H22O12 | + | FORMULA C23H22O12 |
| − | EXACTMASS 490.111126168 | + | EXACTMASS 490.111126168 |
| − | AVERAGEMASS 490.41358 | + | AVERAGEMASS 490.41358 |
| − | SMILES c(c4)(c(OC)cc(c4)C(O3)=CC(c(c23)c(cc(c2)OC(O1)C(O)C(C(O)C1C(O)=O)O)O)=O)OC | + | SMILES c(c4)(c(OC)cc(c4)C(O3)=CC(c(c23)c(cc(c2)OC(O1)C(O)C(C(O)C1C(O)=O)O)O)=O)OC |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
35 38 0 0 0 0 0 0 0 0999 V2000
-1.2980 -0.8973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2980 -1.7067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5969 -2.1116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1042 -1.7067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1042 -0.8973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5969 -0.4925 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8052 -2.1116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5063 -1.7067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5063 -0.8973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8052 -0.4925 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8052 -2.9073 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2071 -0.4927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9216 -0.9052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6361 -0.4927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6361 0.3324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9216 0.7449 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2071 0.3324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5969 -2.9193 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9975 -0.4933 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9868 -0.6057 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4259 -1.3462 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6181 -1.0321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8386 -1.0237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4049 -0.4571 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2302 -0.7534 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.6333 -0.8249 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.0515 -1.2664 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8650 -1.5423 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9216 1.5095 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5758 2.9193 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.8959 -0.1238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.7933 -0.1238 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.4455 0.6563 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2391 0.6806 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.7933 0.6806 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
20 21 1 1 0 0 0
21 22 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 20 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 19 1 0 0 0 0
29 30 1 0 0 0 0
16 29 1 0 0 0 0
31 32 2 0 0 0 0
31 33 1 0 0 0 0
25 31 1 0 0 0 0
34 35 1 0 0 0 0
15 34 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 29 30
M SBL 1 1 33
M SMT 1 OCH3
M SBV 1 33 0.0000 -0.7645
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 3 31 32 33
M SBL 2 1 36
M SMT 2 ^ COOH
M SBV 2 36 0.6657 -0.6296
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 34 35
M SBL 3 1 38
M SMT 3 OCH3
M SBV 3 38 -0.6030 -0.3482
S SKP 5
ID FL3FAFGS0002
FORMULA C23H22O12
EXACTMASS 490.111126168
AVERAGEMASS 490.41358
SMILES c(c4)(c(OC)cc(c4)C(O3)=CC(c(c23)c(cc(c2)OC(O1)C(O)C(C(O)C1C(O)=O)O)O)=O)OC
M END