Mol:FL3FAECS0005
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.4348 -1.0724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4348 -1.0724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4348 -1.7148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4348 -1.7148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8785 -2.0360 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8785 -2.0360 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3222 -1.7148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3222 -1.7148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3222 -1.0724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3222 -1.0724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8785 -0.7513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8785 -0.7513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2341 -2.0360 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2341 -2.0360 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7904 -1.7148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7904 -1.7148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7904 -1.0724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7904 -1.0724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2341 -0.7513 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2341 -0.7513 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2341 -2.5368 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2341 -2.5368 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9909 -0.7514 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9909 -0.7514 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6537 1.7270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6537 1.7270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2593 1.2682 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2593 1.2682 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0024 0.6075 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0024 0.6075 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9955 -0.0299 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9955 -0.0299 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5322 0.4333 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5322 0.4333 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8373 1.0113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8373 1.0113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9086 2.6781 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9086 2.6781 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2941 1.9834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2941 1.9834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6379 0.2290 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6379 0.2290 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8785 -2.6781 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8785 -2.6781 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4702 -0.6482 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4702 -0.6482 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0564 -0.9867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0564 -0.9867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6427 -0.6482 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6427 -0.6482 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6427 0.0287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6427 0.0287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0564 0.3671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0564 0.3671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4702 0.0287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4702 0.0287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4292 -1.7560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4292 -1.7560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0580 -2.2460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0580 -2.2460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5235 -2.0381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5235 -2.0381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0077 -2.0326 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0077 -2.0326 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3825 -1.6577 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3825 -1.6577 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8501 -1.9045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8501 -1.9045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9430 -2.0527 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9430 -2.0527 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6366 -2.2742 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6366 -2.2742 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2172 -2.5523 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2172 -2.5523 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2285 -0.9865 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2285 -0.9865 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2486 -1.1203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2486 -1.1203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4517 -1.3339 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4517 -1.3339 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1414 1.4130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1414 1.4130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5731 1.0005 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5731 1.0005 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2285 0.3669 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2285 0.3669 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9430 -0.0456 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9430 -0.0456 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 1 12 1 0 0 0 0 | + | 1 12 1 0 0 0 0 |
| − | 13 14 1 1 0 0 0 | + | 13 14 1 1 0 0 0 |
| − | 14 15 1 1 0 0 0 | + | 14 15 1 1 0 0 0 |
| − | 16 15 1 1 0 0 0 | + | 16 15 1 1 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 13 19 1 0 0 0 0 | + | 13 19 1 0 0 0 0 |
| − | 14 20 1 0 0 0 0 | + | 14 20 1 0 0 0 0 |
| − | 15 21 1 0 0 0 0 | + | 15 21 1 0 0 0 0 |
| − | 6 16 1 0 0 0 0 | + | 6 16 1 0 0 0 0 |
| − | 3 22 1 0 0 0 0 | + | 3 22 1 0 0 0 0 |
| − | 23 24 2 0 0 0 0 | + | 23 24 2 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 2 0 0 0 0 | + | 25 26 2 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 2 0 0 0 0 | + | 27 28 2 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 9 23 1 0 0 0 0 | + | 9 23 1 0 0 0 0 |
| − | 29 30 1 1 0 0 0 | + | 29 30 1 1 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 29 1 0 0 0 0 | + | 34 29 1 0 0 0 0 |
| − | 29 35 1 0 0 0 0 | + | 29 35 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 2 1 0 0 0 0 | + | 32 2 1 0 0 0 0 |
| − | 25 38 1 0 0 0 0 | + | 25 38 1 0 0 0 0 |
| − | 34 39 1 0 0 0 0 | + | 34 39 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 18 41 1 0 0 0 0 | + | 18 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 26 43 1 0 0 0 0 | + | 26 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 39 40 | + | M SAL 1 2 39 40 |
| − | M SBL 1 1 43 | + | M SBL 1 1 43 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 43 -6.8823 6.3118 | + | M SBV 1 43 -6.8823 6.3118 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 41 42 | + | M SAL 2 2 41 42 |
| − | M SBL 2 1 45 | + | M SBL 2 1 45 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 45 -5.7880 5.9293 | + | M SBV 2 45 -5.7880 5.9293 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 43 44 | + | M SAL 3 2 43 44 |
| − | M SBL 3 1 47 | + | M SBL 3 1 47 |
| − | M SMT 3 OCH3 | + | M SMT 3 OCH3 |
| − | M SBV 3 47 -5.8980 5.8658 | + | M SBV 3 47 -5.8980 5.8658 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FAECS0005 | + | ID FL3FAECS0005 |
| − | KNApSAcK_ID C00006284 | + | KNApSAcK_ID C00006284 |
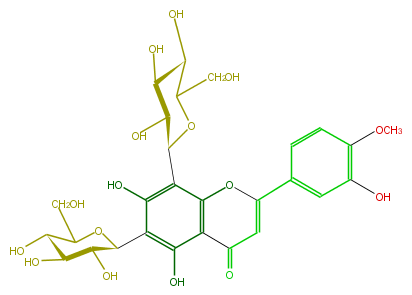
| − | NAME 6,8-Di-C-glucopyranosyldiosmetin | + | NAME 6,8-Di-C-glucopyranosyldiosmetin |
| − | CAS_RN 98813-28-6 | + | CAS_RN 98813-28-6 |
| − | FORMULA C28H32O16 | + | FORMULA C28H32O16 |
| − | EXACTMASS 624.1690349759999 | + | EXACTMASS 624.1690349759999 |
| − | AVERAGEMASS 624.54408 | + | AVERAGEMASS 624.54408 |
| − | SMILES OC(C(O)1)C(C(OC1c(c53)c(O)c(c(c3C(C=C(O5)c(c4)ccc(OC)c4O)=O)O)C(C(O)2)OC(C(C2O)O)CO)CO)O | + | SMILES OC(C(O)1)C(C(OC1c(c53)c(O)c(c(c3C(C=C(O5)c(c4)ccc(OC)c4O)=O)O)C(C(O)2)OC(C(C2O)O)CO)CO)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
-1.4348 -1.0724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4348 -1.7148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8785 -2.0360 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3222 -1.7148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3222 -1.0724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8785 -0.7513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2341 -2.0360 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7904 -1.7148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7904 -1.0724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2341 -0.7513 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2341 -2.5368 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9909 -0.7514 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6537 1.7270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2593 1.2682 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0024 0.6075 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9955 -0.0299 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5322 0.4333 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8373 1.0113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9086 2.6781 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2941 1.9834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6379 0.2290 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8785 -2.6781 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4702 -0.6482 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0564 -0.9867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6427 -0.6482 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6427 0.0287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0564 0.3671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4702 0.0287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4292 -1.7560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0580 -2.2460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5235 -2.0381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0077 -2.0326 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3825 -1.6577 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8501 -1.9045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9430 -2.0527 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6366 -2.2742 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2172 -2.5523 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2285 -0.9865 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2486 -1.1203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4517 -1.3339 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1414 1.4130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5731 1.0005 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2285 0.3669 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9430 -0.0456 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
1 12 1 0 0 0 0
13 14 1 1 0 0 0
14 15 1 1 0 0 0
16 15 1 1 0 0 0
16 17 1 0 0 0 0
17 18 1 0 0 0 0
18 13 1 0 0 0 0
13 19 1 0 0 0 0
14 20 1 0 0 0 0
15 21 1 0 0 0 0
6 16 1 0 0 0 0
3 22 1 0 0 0 0
23 24 2 0 0 0 0
24 25 1 0 0 0 0
25 26 2 0 0 0 0
26 27 1 0 0 0 0
27 28 2 0 0 0 0
28 23 1 0 0 0 0
9 23 1 0 0 0 0
29 30 1 1 0 0 0
30 31 1 1 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 29 1 0 0 0 0
29 35 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 2 1 0 0 0 0
25 38 1 0 0 0 0
34 39 1 0 0 0 0
39 40 1 0 0 0 0
18 41 1 0 0 0 0
41 42 1 0 0 0 0
26 43 1 0 0 0 0
43 44 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 39 40
M SBL 1 1 43
M SMT 1 ^CH2OH
M SBV 1 43 -6.8823 6.3118
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 41 42
M SBL 2 1 45
M SMT 2 CH2OH
M SBV 2 45 -5.7880 5.9293
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 43 44
M SBL 3 1 47
M SMT 3 OCH3
M SBV 3 47 -5.8980 5.8658
S SKP 8
ID FL3FAECS0005
KNApSAcK_ID C00006284
NAME 6,8-Di-C-glucopyranosyldiosmetin
CAS_RN 98813-28-6
FORMULA C28H32O16
EXACTMASS 624.1690349759999
AVERAGEMASS 624.54408
SMILES OC(C(O)1)C(C(OC1c(c53)c(O)c(c(c3C(C=C(O5)c(c4)ccc(OC)c4O)=O)O)C(C(O)2)OC(C(C2O)O)CO)CO)O
M END