Mol:FL3FADGS0017
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 55 60 0 0 0 0 0 0 0 0999 V2000 | + | 55 60 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.3859 1.5973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3859 1.5973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3859 1.0765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3859 1.0765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8370 0.8161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8370 0.8161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2880 1.0765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2880 1.0765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2880 1.5973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2880 1.5973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8370 1.8577 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8370 1.8577 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7391 0.8161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7391 0.8161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1902 1.0765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1902 1.0765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1902 1.5973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1902 1.5973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7391 1.8577 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7391 1.8577 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9108 0.4482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9108 0.4482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6411 1.8577 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6411 1.8577 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1008 1.5922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1008 1.5922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5605 1.8577 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5605 1.8577 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5605 2.3885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5605 2.3885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1008 2.6539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1008 2.6539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6411 2.3885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6411 2.3885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5605 2.3885 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5605 2.3885 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0642 1.8572 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0642 1.8572 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7037 1.6693 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -2.7037 1.6693 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -2.1880 0.9887 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.1880 0.9887 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.4455 1.2775 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.4455 1.2775 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.7291 1.2852 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.7291 1.2852 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.2497 1.8059 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2497 1.8059 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8993 1.4631 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -1.8993 1.4631 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -3.4175 1.2572 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4175 1.2572 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9918 0.9496 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9918 0.9496 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0201 0.5633 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0201 0.5633 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8370 0.2963 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8370 0.2963 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1137 -0.2574 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -3.1137 -0.2574 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -2.5981 -0.9380 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.5981 -0.9380 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.8556 -0.6493 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.8556 -0.6493 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.1391 -0.6416 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.1391 -0.6416 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.6597 -0.1208 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6597 -0.1208 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4182 -0.3932 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -2.4182 -0.3932 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -3.6902 -0.5903 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6902 -0.5903 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3100 -1.3131 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3100 -1.3131 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1681 -1.1168 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1681 -1.1168 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5983 -4.0858 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5983 -4.0858 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9634 -3.1706 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -0.9634 -3.1706 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -0.1724 -2.8488 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.1724 -2.8488 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.2592 -2.0568 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.2592 -2.0568 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.0811 -1.3630 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.0811 -1.3630 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.7190 -1.7308 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7190 -1.7308 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6522 -2.5340 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -0.6522 -2.5340 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -0.3342 -3.4526 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3342 -3.4526 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5409 -1.8309 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5409 -1.8309 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3306 3.1991 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3306 3.1991 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7190 4.1206 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7190 4.1206 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5171 -2.5033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5171 -2.5033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4377 -2.8936 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4377 -2.8936 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1002 2.4038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1002 2.4038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0516 2.7119 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0516 2.7119 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0272 0.2137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0272 0.2137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8396 0.3570 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8396 0.3570 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 18 15 1 0 0 0 0 | + | 18 15 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 19 1 0 0 0 0 | + | 23 19 1 0 0 0 0 |
| − | 3 29 1 0 0 0 0 | + | 3 29 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 40 41 1 1 0 0 0 | + | 40 41 1 1 0 0 0 |
| − | 41 42 1 1 0 0 0 | + | 41 42 1 1 0 0 0 |
| − | 43 42 1 1 0 0 0 | + | 43 42 1 1 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 40 1 0 0 0 0 | + | 45 40 1 0 0 0 0 |
| − | 41 46 1 0 0 0 0 | + | 41 46 1 0 0 0 0 |
| − | 42 47 1 0 0 0 0 | + | 42 47 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 43 38 1 0 0 0 0 | + | 43 38 1 0 0 0 0 |
| − | 33 28 1 0 0 0 0 | + | 33 28 1 0 0 0 0 |
| − | 16 48 1 0 0 0 0 | + | 16 48 1 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 45 50 1 0 0 0 0 | + | 45 50 1 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 25 52 1 0 0 0 0 | + | 25 52 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 35 54 1 0 0 0 0 | + | 35 54 1 0 0 0 0 |
| − | 54 55 1 0 0 0 0 | + | 54 55 1 0 0 0 0 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 54 55 | + | M SAL 4 2 54 55 |
| − | M SBL 4 1 59 | + | M SBL 4 1 59 |
| − | M SMT 4 CH2OH | + | M SMT 4 CH2OH |
| − | M SVB 4 59 -2.7137 0.4142 | + | M SVB 4 59 -2.7137 0.4142 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 52 53 | + | M SAL 3 2 52 53 |
| − | M SBL 3 1 57 | + | M SBL 3 1 57 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SVB 3 57 -2.1002 2.4038 | + | M SVB 3 57 -2.1002 2.4038 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 50 51 | + | M SAL 2 2 50 51 |
| − | M SBL 2 1 55 | + | M SBL 2 1 55 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 55 -2.86 -1.3854 | + | M SVB 2 55 -2.86 -1.3854 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 48 49 | + | M SAL 1 2 48 49 |
| − | M SBL 1 1 53 | + | M SBL 1 1 53 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 53 3.3306 3.1991 | + | M SVB 1 53 3.3306 3.1991 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FADGS0017 | + | ID FL3FADGS0017 |
| − | KNApSAcK_ID C00004351 | + | KNApSAcK_ID C00004351 |
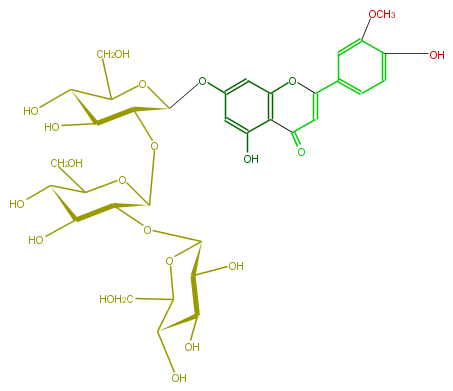
| − | NAME Luteolin 3'-methyl ether 7-sophorotrioside | + | NAME Luteolin 3'-methyl ether 7-sophorotrioside |
| − | CAS_RN 63069-54-5 | + | CAS_RN 63069-54-5 |
| − | FORMULA C34H42O21 | + | FORMULA C34H42O21 |
| − | EXACTMASS 786.2218584059999 | + | EXACTMASS 786.2218584059999 |
| − | AVERAGEMASS 786.68468 | + | AVERAGEMASS 786.68468 |
| − | SMILES C(C(O3)[C@H](O)[C@@H]([C@@H]([C@H](Oc(c6)cc(c(c65)C(=O)C=C(O5)c(c4)cc(c(O)c4)OC)O)3)O[C@@H]([C@@H](O[C@@H]([C@@H](O)2)OC(CO)[C@@H]([C@@H]2O)O)1)OC([C@H](O)[C@@H]1O)CO)O)O | + | SMILES C(C(O3)[C@H](O)[C@@H]([C@@H]([C@H](Oc(c6)cc(c(c65)C(=O)C=C(O5)c(c4)cc(c(O)c4)OC)O)3)O[C@@H]([C@@H](O[C@@H]([C@@H](O)2)OC(CO)[C@@H]([C@@H]2O)O)1)OC([C@H](O)[C@@H]1O)CO)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
55 60 0 0 0 0 0 0 0 0999 V2000
0.3859 1.5973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3859 1.0765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8370 0.8161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2880 1.0765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2880 1.5973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8370 1.8577 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7391 0.8161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1902 1.0765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1902 1.5973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7391 1.8577 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9108 0.4482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6411 1.8577 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1008 1.5922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5605 1.8577 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5605 2.3885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1008 2.6539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6411 2.3885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5605 2.3885 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0642 1.8572 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7037 1.6693 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-2.1880 0.9887 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.4455 1.2775 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.7291 1.2852 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.2497 1.8059 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8993 1.4631 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.4175 1.2572 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9918 0.9496 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0201 0.5633 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8370 0.2963 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1137 -0.2574 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-2.5981 -0.9380 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.8556 -0.6493 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.1391 -0.6416 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.6597 -0.1208 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4182 -0.3932 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.6902 -0.5903 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3100 -1.3131 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1681 -1.1168 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5983 -4.0858 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9634 -3.1706 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-0.1724 -2.8488 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.2592 -2.0568 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.0811 -1.3630 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.7190 -1.7308 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6522 -2.5340 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-0.3342 -3.4526 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5409 -1.8309 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3306 3.1991 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7190 4.1206 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5171 -2.5033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4377 -2.8936 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1002 2.4038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0516 2.7119 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0272 0.2137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8396 0.3570 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
18 15 1 0 0 0 0
1 19 1 0 0 0 0
20 21 1 1 0 0 0
21 22 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 20 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 19 1 0 0 0 0
3 29 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
40 41 1 1 0 0 0
41 42 1 1 0 0 0
43 42 1 1 0 0 0
43 44 1 0 0 0 0
44 45 1 0 0 0 0
45 40 1 0 0 0 0
41 46 1 0 0 0 0
42 47 1 0 0 0 0
39 40 1 0 0 0 0
43 38 1 0 0 0 0
33 28 1 0 0 0 0
16 48 1 0 0 0 0
48 49 1 0 0 0 0
45 50 1 0 0 0 0
50 51 1 0 0 0 0
25 52 1 0 0 0 0
52 53 1 0 0 0 0
35 54 1 0 0 0 0
54 55 1 0 0 0 0
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 54 55
M SBL 4 1 59
M SMT 4 CH2OH
M SVB 4 59 -2.7137 0.4142
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 52 53
M SBL 3 1 57
M SMT 3 CH2OH
M SVB 3 57 -2.1002 2.4038
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 50 51
M SBL 2 1 55
M SMT 2 CH2OH
M SVB 2 55 -2.86 -1.3854
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 48 49
M SBL 1 1 53
M SMT 1 OCH3
M SVB 1 53 3.3306 3.1991
S SKP 8
ID FL3FADGS0017
KNApSAcK_ID C00004351
NAME Luteolin 3'-methyl ether 7-sophorotrioside
CAS_RN 63069-54-5
FORMULA C34H42O21
EXACTMASS 786.2218584059999
AVERAGEMASS 786.68468
SMILES C(C(O3)[C@H](O)[C@@H]([C@@H]([C@H](Oc(c6)cc(c(c65)C(=O)C=C(O5)c(c4)cc(c(O)c4)OC)O)3)O[C@@H]([C@@H](O[C@@H]([C@@H](O)2)OC(CO)[C@@H]([C@@H]2O)O)1)OC([C@H](O)[C@@H]1O)CO)O)O
M END