Mol:FL3FACCS0056
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | 1.9761 1.8615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9761 1.8615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2617 2.2740 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2617 2.2740 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5472 1.8615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5472 1.8615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5472 1.0365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5472 1.0365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2617 0.6240 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2617 0.6240 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9761 1.0365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9761 1.0365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1673 2.2740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1673 2.2740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8818 1.8615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8818 1.8615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8818 1.0365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8818 1.0365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1673 0.6240 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1673 0.6240 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2617 0.0519 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2617 0.0519 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7775 2.3242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7775 2.3242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4920 1.9117 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4920 1.9117 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2064 2.3242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2064 2.3242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2064 3.1492 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2064 3.1492 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4920 3.5617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4920 3.5617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7775 3.1492 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7775 3.1492 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8899 3.5438 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8899 3.5438 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1673 -0.1460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1673 -0.1460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5722 2.2601 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5722 2.2601 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8680 1.9422 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8680 1.9422 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7350 1.4000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7350 1.4000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3224 0.6853 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3224 0.6853 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5241 0.8944 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5241 0.8944 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7306 0.6676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7306 0.6676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1432 1.3823 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1432 1.3823 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9416 1.1733 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9416 1.1733 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0985 1.7591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0985 1.7591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5784 2.0361 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5784 2.0361 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2182 0.9168 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2182 0.9168 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7711 0.9443 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7711 0.9443 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7448 -1.5018 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7448 -1.5018 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4584 -1.0878 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4584 -1.0878 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1738 -1.4988 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1738 -1.4988 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1755 -2.3238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1755 -2.3238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4619 -2.7378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4619 -2.7378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7466 -2.3268 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7466 -2.3268 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0339 -2.7402 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0339 -2.7402 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4637 -3.5617 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4637 -3.5617 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8899 -2.7342 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8899 -2.7342 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4567 -0.2639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4567 -0.2639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1694 0.1495 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1694 0.1495 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7423 0.1465 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7423 0.1465 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 3 7 1 0 0 0 0 | + | 3 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 4 1 0 0 0 0 | + | 10 4 1 0 0 0 0 |
| − | 5 11 2 0 0 0 0 | + | 5 11 2 0 0 0 0 |
| − | 1 12 1 0 0 0 0 | + | 1 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 15 18 1 0 0 0 0 | + | 15 18 1 0 0 0 0 |
| − | 10 19 1 0 0 0 0 | + | 10 19 1 0 0 0 0 |
| − | 8 20 1 0 0 0 0 | + | 8 20 1 0 0 0 0 |
| − | 14 21 1 0 0 0 0 | + | 14 21 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 22 30 1 0 0 0 0 | + | 22 30 1 0 0 0 0 |
| − | 23 31 1 0 0 0 0 | + | 23 31 1 0 0 0 0 |
| − | 25 9 1 0 0 0 0 | + | 25 9 1 0 0 0 0 |
| − | 32 33 2 0 0 0 0 | + | 32 33 2 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 2 0 0 0 0 | + | 34 35 2 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 36 39 1 0 0 0 0 | + | 36 39 1 0 0 0 0 |
| − | 35 40 1 0 0 0 0 | + | 35 40 1 0 0 0 0 |
| − | 33 41 1 0 0 0 0 | + | 33 41 1 0 0 0 0 |
| − | 41 42 2 0 0 0 0 | + | 41 42 2 0 0 0 0 |
| − | 41 43 1 0 0 0 0 | + | 41 43 1 0 0 0 0 |
| − | 43 24 1 0 0 0 0 | + | 43 24 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FACCS0056 | + | ID FL3FACCS0056 |
| − | KNApSAcK_ID C00014098 | + | KNApSAcK_ID C00014098 |
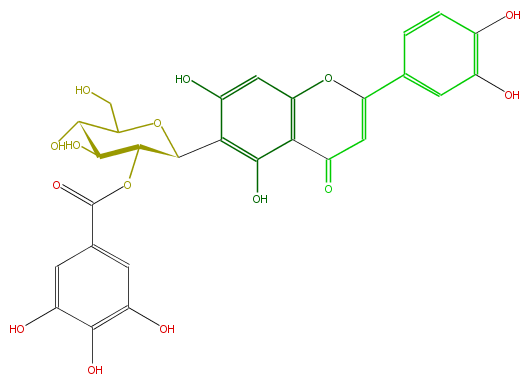
| − | NAME Isoorientin 2''-O-gallate | + | NAME Isoorientin 2''-O-gallate |
| − | CAS_RN 438000-11-4 | + | CAS_RN 438000-11-4 |
| − | FORMULA C28H24O15 | + | FORMULA C28H24O15 |
| − | EXACTMASS 600.111520098 | + | EXACTMASS 600.111520098 |
| − | AVERAGEMASS 600.48116 | + | AVERAGEMASS 600.48116 |
| − | SMILES c(c5O)(ccc(c5)C(O1)=CC(=O)c(c(O)2)c1cc(O)c(C(O3)C(OC(c(c4)cc(O)c(O)c4O)=O)C(O)C(C(CO)3)O)2)O | + | SMILES c(c5O)(ccc(c5)C(O1)=CC(=O)c(c(O)2)c1cc(O)c(C(O3)C(OC(c(c4)cc(O)c(O)c4O)=O)C(O)C(C(CO)3)O)2)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
1.9761 1.8615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2617 2.2740 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5472 1.8615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5472 1.0365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2617 0.6240 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9761 1.0365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1673 2.2740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8818 1.8615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8818 1.0365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1673 0.6240 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2617 0.0519 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7775 2.3242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4920 1.9117 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2064 2.3242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2064 3.1492 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4920 3.5617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7775 3.1492 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8899 3.5438 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1673 -0.1460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5722 2.2601 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.8680 1.9422 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7350 1.4000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3224 0.6853 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5241 0.8944 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7306 0.6676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1432 1.3823 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9416 1.1733 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0985 1.7591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5784 2.0361 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2182 0.9168 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7711 0.9443 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7448 -1.5018 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4584 -1.0878 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1738 -1.4988 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1755 -2.3238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4619 -2.7378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7466 -2.3268 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0339 -2.7402 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4637 -3.5617 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8899 -2.7342 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4567 -0.2639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1694 0.1495 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7423 0.1465 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
3 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 4 1 0 0 0 0
5 11 2 0 0 0 0
1 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
15 18 1 0 0 0 0
10 19 1 0 0 0 0
8 20 1 0 0 0 0
14 21 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
27 28 1 0 0 0 0
28 29 1 0 0 0 0
22 30 1 0 0 0 0
23 31 1 0 0 0 0
25 9 1 0 0 0 0
32 33 2 0 0 0 0
33 34 1 0 0 0 0
34 35 2 0 0 0 0
35 36 1 0 0 0 0
36 37 2 0 0 0 0
37 32 1 0 0 0 0
37 38 1 0 0 0 0
36 39 1 0 0 0 0
35 40 1 0 0 0 0
33 41 1 0 0 0 0
41 42 2 0 0 0 0
41 43 1 0 0 0 0
43 24 1 0 0 0 0
S SKP 8
ID FL3FACCS0056
KNApSAcK_ID C00014098
NAME Isoorientin 2''-O-gallate
CAS_RN 438000-11-4
FORMULA C28H24O15
EXACTMASS 600.111520098
AVERAGEMASS 600.48116
SMILES c(c5O)(ccc(c5)C(O1)=CC(=O)c(c(O)2)c1cc(O)c(C(O3)C(OC(c(c4)cc(O)c(O)c4O)=O)C(O)C(C(CO)3)O)2)O
M END