Mol:FL3FACCS0047
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.6182 1.0973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6182 1.0973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0484 1.4821 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0484 1.4821 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0484 2.2518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0484 2.2518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6182 2.6367 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6182 2.6367 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2847 2.2518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2847 2.2518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2847 1.4821 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2847 1.4821 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7150 1.0973 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7150 1.0973 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3815 1.4821 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3815 1.4821 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3815 2.2518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3815 2.2518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7150 2.6367 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7150 2.6367 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0199 1.1136 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0199 1.1136 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0199 0.3829 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0199 0.3829 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6527 0.0176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6527 0.0176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2855 0.3829 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2855 0.3829 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2855 1.1136 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2855 1.1136 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6527 1.4789 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6527 1.4789 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7201 0.1320 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7201 0.1320 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7150 3.0942 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7150 3.0942 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6182 3.0965 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6182 3.0965 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8124 1.1775 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8124 1.1775 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2941 0.4043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2941 0.4043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7543 -0.2196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7543 -0.2196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0099 0.1362 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0099 0.1362 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1882 0.0630 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1882 0.0630 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7279 0.6870 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7279 0.6870 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4724 0.3313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4724 0.3313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7960 -0.0976 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7960 -0.0976 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3688 -0.0549 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3688 -0.0549 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6943 -0.4105 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6943 -0.4105 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7953 0.8906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7953 0.8906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1955 1.1217 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1955 1.1217 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3208 -2.0320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3208 -2.0320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9365 -2.7620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9365 -2.7620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7052 -1.9701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7052 -1.9701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1025 -1.4067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1025 -1.4067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4868 -0.6766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4868 -0.6766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7182 -1.4685 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7182 -1.4685 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8062 -3.0965 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8062 -3.0965 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0807 -0.8480 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0807 -0.8480 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3426 -2.5898 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3426 -2.5898 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7293 -2.7405 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7293 -2.7405 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1879 -2.1973 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1879 -2.1973 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7960 1.4083 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7960 1.4083 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 2 7 1 0 0 0 0 | + | 2 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 3 1 0 0 0 0 | + | 10 3 1 0 0 0 0 |
| − | 8 11 1 0 0 0 0 | + | 8 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 14 17 1 0 0 0 0 | + | 14 17 1 0 0 0 0 |
| − | 10 18 2 0 0 0 0 | + | 10 18 2 0 0 0 0 |
| − | 4 19 1 0 0 0 0 | + | 4 19 1 0 0 0 0 |
| − | 6 20 1 0 0 0 0 | + | 6 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 1 24 1 0 0 0 0 | + | 1 24 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 36 35 1 1 0 0 0 | + | 36 35 1 1 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 33 38 1 0 0 0 0 | + | 33 38 1 0 0 0 0 |
| − | 35 39 1 0 0 0 0 | + | 35 39 1 0 0 0 0 |
| − | 32 40 1 0 0 0 0 | + | 32 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 36 29 1 0 0 0 0 | + | 36 29 1 0 0 0 0 |
| − | 34 42 1 0 0 0 0 | + | 34 42 1 0 0 0 0 |
| − | 15 43 1 0 0 0 0 | + | 15 43 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
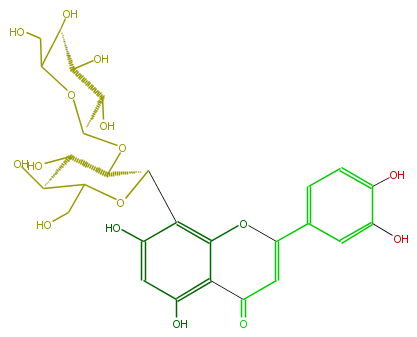
| − | ID FL3FACCS0047 | + | ID FL3FACCS0047 |
| − | KNApSAcK_ID C00011124 | + | KNApSAcK_ID C00011124 |
| − | NAME 2''-O-beta-L-galactopyranosylorientin | + | NAME 2''-O-beta-L-galactopyranosylorientin |
| − | CAS_RN 861691-37-4 | + | CAS_RN 861691-37-4 |
| − | FORMULA C27H30O16 | + | FORMULA C27H30O16 |
| − | EXACTMASS 610.153384912 | + | EXACTMASS 610.153384912 |
| − | AVERAGEMASS 610.5175 | + | AVERAGEMASS 610.5175 |
| − | SMILES C(C(C1O)OC(OC(C(O)2)C(c(c34)c(cc(c3C(C=C(c(c5)ccc(O)c5O)O4)=O)O)O)OC(C2O)CO)C(O)C(O)1)O | + | SMILES C(C(C1O)OC(OC(C(O)2)C(c(c34)c(cc(c3C(C=C(c(c5)ccc(O)c5O)O4)=O)O)O)OC(C2O)CO)C(O)C(O)1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-0.6182 1.0973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0484 1.4821 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0484 2.2518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6182 2.6367 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2847 2.2518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2847 1.4821 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7150 1.0973 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3815 1.4821 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3815 2.2518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7150 2.6367 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0199 1.1136 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0199 0.3829 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6527 0.0176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2855 0.3829 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2855 1.1136 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6527 1.4789 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7201 0.1320 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7150 3.0942 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6182 3.0965 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8124 1.1775 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2941 0.4043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7543 -0.2196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0099 0.1362 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1882 0.0630 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7279 0.6870 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4724 0.3313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7960 -0.0976 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3688 -0.0549 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6943 -0.4105 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7953 0.8906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1955 1.1217 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3208 -2.0320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9365 -2.7620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7052 -1.9701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1025 -1.4067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4868 -0.6766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7182 -1.4685 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8062 -3.0965 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0807 -0.8480 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3426 -2.5898 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7293 -2.7405 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1879 -2.1973 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7960 1.4083 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
2 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 3 1 0 0 0 0
8 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
14 17 1 0 0 0 0
10 18 2 0 0 0 0
4 19 1 0 0 0 0
6 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
1 24 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
26 30 1 0 0 0 0
30 31 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
36 35 1 1 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
33 38 1 0 0 0 0
35 39 1 0 0 0 0
32 40 1 0 0 0 0
40 41 1 0 0 0 0
36 29 1 0 0 0 0
34 42 1 0 0 0 0
15 43 1 0 0 0 0
S SKP 8
ID FL3FACCS0047
KNApSAcK_ID C00011124
NAME 2''-O-beta-L-galactopyranosylorientin
CAS_RN 861691-37-4
FORMULA C27H30O16
EXACTMASS 610.153384912
AVERAGEMASS 610.5175
SMILES C(C(C1O)OC(OC(C(O)2)C(c(c34)c(cc(c3C(C=C(c(c5)ccc(O)c5O)O4)=O)O)O)OC(C2O)CO)C(O)C(O)1)O
M END