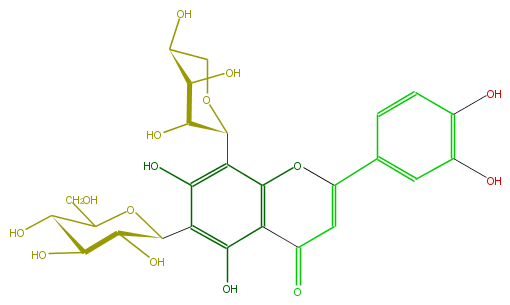
Mol:FL3FACCS0022
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 41 45 0 0 0 0 0 0 0 0999 V2000 | + | 41 45 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.2613 -0.6639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2613 -0.6639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2613 -1.4890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2613 -1.4890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5468 -1.9015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5468 -1.9015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1676 -1.4890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1676 -1.4890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1676 -0.6639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1676 -0.6639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5468 -0.2514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5468 -0.2514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8821 -1.9015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8821 -1.9015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5966 -1.4890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5966 -1.4890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5966 -0.6639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5966 -0.6639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8821 -0.2514 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8821 -0.2514 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8821 -2.7828 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8821 -2.7828 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5468 -2.7261 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5468 -2.7261 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4696 -0.1191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4696 -0.1191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2225 -0.5539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2225 -0.5539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9755 -0.1191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9755 -0.1191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9755 0.7502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9755 0.7502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2225 1.1848 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2225 1.1848 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4696 0.7502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4696 0.7502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7278 1.1846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7278 1.1846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9921 -0.2553 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9921 -0.2553 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0579 -1.2696 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0579 -1.2696 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3957 -2.0113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3957 -2.0113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7336 -1.6139 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7336 -1.6139 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8329 -1.7198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8329 -1.7198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4686 -1.1372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4686 -1.1372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1838 -1.4815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1838 -1.4815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6733 -1.6055 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6733 -1.6055 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2194 -2.0361 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2194 -2.0361 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0048 -2.1798 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0048 -2.1798 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3080 0.5813 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3080 0.5813 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5411 0.3758 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5411 0.3758 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9493 1.0567 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9493 1.0567 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9375 1.8551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9375 1.8551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7044 2.0607 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7044 2.0607 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2961 1.3797 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2961 1.3797 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4816 1.5979 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4816 1.5979 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9181 0.3648 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9181 0.3648 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4655 2.7828 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4655 2.7828 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7278 -0.5536 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7278 -0.5536 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7044 -0.9313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7044 -0.9313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7278 -0.6570 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7278 -0.6570 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 3 12 1 0 0 0 0 | + | 3 12 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 2 0 0 0 0 | + | 17 18 2 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 9 13 1 0 0 0 0 | + | 9 13 1 0 0 0 0 |
| − | 16 19 1 0 0 0 0 | + | 16 19 1 0 0 0 0 |
| − | 1 20 1 0 0 0 0 | + | 1 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 2 1 0 0 0 0 | + | 24 2 1 0 0 0 0 |
| − | 31 30 1 1 0 0 0 | + | 31 30 1 1 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 35 30 1 1 0 0 0 | + | 35 30 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 30 37 1 0 0 0 0 | + | 30 37 1 0 0 0 0 |
| − | 6 31 1 0 0 0 0 | + | 6 31 1 0 0 0 0 |
| − | 34 38 1 0 0 0 0 | + | 34 38 1 0 0 0 0 |
| − | 15 39 1 0 0 0 0 | + | 15 39 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 26 40 1 0 0 0 0 | + | 26 40 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 45 0.5205 -0.5502 | + | M SBV 1 45 0.5205 -0.5502 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FACCS0022 | + | ID FL3FACCS0022 |
| − | FORMULA C26H28O15 | + | FORMULA C26H28O15 |
| − | EXACTMASS 580.1428202259999 | + | EXACTMASS 580.1428202259999 |
| − | AVERAGEMASS 580.49152 | + | AVERAGEMASS 580.49152 |
| − | SMILES C(C(C1O)OC(c(c4O)c(c(c(c4C(O5)C(O)C(O)C(O)C5)2)C(C=C(c(c3)ccc(O)c3O)O2)=O)O)C(C1O)O)O | + | SMILES C(C(C1O)OC(c(c4O)c(c(c(c4C(O5)C(O)C(O)C(O)C5)2)C(C=C(c(c3)ccc(O)c3O)O2)=O)O)C(C1O)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
41 45 0 0 0 0 0 0 0 0999 V2000
-1.2613 -0.6639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2613 -1.4890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5468 -1.9015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1676 -1.4890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1676 -0.6639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5468 -0.2514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8821 -1.9015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5966 -1.4890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5966 -0.6639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8821 -0.2514 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8821 -2.7828 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5468 -2.7261 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4696 -0.1191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2225 -0.5539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9755 -0.1191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9755 0.7502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2225 1.1848 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4696 0.7502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7278 1.1846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9921 -0.2553 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0579 -1.2696 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3957 -2.0113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7336 -1.6139 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8329 -1.7198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4686 -1.1372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1838 -1.4815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6733 -1.6055 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2194 -2.0361 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0048 -2.1798 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3080 0.5813 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5411 0.3758 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9493 1.0567 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9375 1.8551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7044 2.0607 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2961 1.3797 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4816 1.5979 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9181 0.3648 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4655 2.7828 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.7278 -0.5536 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7044 -0.9313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7278 -0.6570 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
3 12 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 17 1 0 0 0 0
17 18 2 0 0 0 0
18 13 1 0 0 0 0
9 13 1 0 0 0 0
16 19 1 0 0 0 0
1 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 2 1 0 0 0 0
31 30 1 1 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 1 0 0 0
35 30 1 1 0 0 0
35 36 1 0 0 0 0
30 37 1 0 0 0 0
6 31 1 0 0 0 0
34 38 1 0 0 0 0
15 39 1 0 0 0 0
40 41 1 0 0 0 0
26 40 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 45
M SMT 1 ^CH2OH
M SBV 1 45 0.5205 -0.5502
S SKP 5
ID FL3FACCS0022
FORMULA C26H28O15
EXACTMASS 580.1428202259999
AVERAGEMASS 580.49152
SMILES C(C(C1O)OC(c(c4O)c(c(c(c4C(O5)C(O)C(O)C(O)C5)2)C(C=C(c(c3)ccc(O)c3O)O2)=O)O)C(C1O)O)O
M END