Mol:FL3FAAGS0065
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.5988 2.0440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5988 2.0440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5988 1.2190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5988 1.2190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1157 0.8065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1157 0.8065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8302 1.2190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8302 1.2190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8302 2.0440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8302 2.0440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1157 2.4565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1157 2.4565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5446 0.8065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5446 0.8065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2591 1.2190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2591 1.2190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2591 2.0440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2591 2.0440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5446 2.4565 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5446 2.4565 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9736 2.4565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9736 2.4565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6881 2.0440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6881 2.0440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4025 2.4565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4025 2.4565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4025 3.2815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4025 3.2815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6881 3.6940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6881 3.6940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9736 3.2815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9736 3.2815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5446 -0.0185 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5446 -0.0185 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1157 -0.0185 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1157 -0.0185 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1488 3.7124 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1488 3.7124 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3184 2.3910 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3184 2.3910 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0279 2.4678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0279 2.4678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6153 1.7531 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6153 1.7531 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8169 1.9623 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8169 1.9623 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0234 1.7354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0234 1.7354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4360 2.4502 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4360 2.4502 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2344 2.2411 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2344 2.2411 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3914 2.8270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3914 2.8270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8712 3.1040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8712 3.1040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5111 1.9846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5111 1.9846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0639 2.0122 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0639 2.0122 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0478 1.1783 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0478 1.1783 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3250 1.2940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3250 1.2940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1488 1.3023 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1488 1.3023 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3328 0.4443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3328 0.4443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3379 -0.3810 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3379 -0.3810 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3417 -1.2386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3417 -1.2386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0600 -1.6444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0600 -1.6444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0678 -2.4694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0678 -2.4694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3572 -2.8886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3572 -2.8886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6389 -2.4828 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6389 -2.4828 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6312 -1.6578 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6312 -1.6578 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3649 -3.7124 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3649 -3.7124 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 7 17 2 0 0 0 0 | + | 7 17 2 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 14 19 1 0 0 0 0 | + | 14 19 1 0 0 0 0 |
| − | 20 1 1 0 0 0 0 | + | 20 1 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 21 29 1 0 0 0 0 | + | 21 29 1 0 0 0 0 |
| − | 22 30 1 0 0 0 0 | + | 22 30 1 0 0 0 0 |
| − | 24 20 1 0 0 0 0 | + | 24 20 1 0 0 0 0 |
| − | 32 33 2 0 0 0 0 | + | 32 33 2 0 0 0 0 |
| − | 32 34 1 0 0 0 0 | + | 32 34 1 0 0 0 0 |
| − | 34 35 2 0 0 0 0 | + | 34 35 2 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 2 0 0 0 0 | + | 40 41 2 0 0 0 0 |
| − | 41 36 1 0 0 0 0 | + | 41 36 1 0 0 0 0 |
| − | 39 42 1 0 0 0 0 | + | 39 42 1 0 0 0 0 |
| − | 31 23 1 0 0 0 0 | + | 31 23 1 0 0 0 0 |
| − | 32 30 1 0 0 0 0 | + | 32 30 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FAAGS0065 | + | ID FL3FAAGS0065 |
| − | KNApSAcK_ID C00013615 | + | KNApSAcK_ID C00013615 |
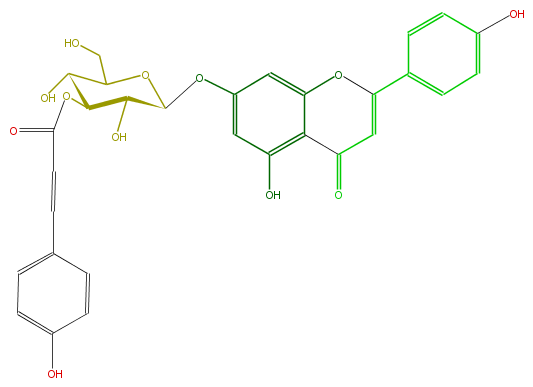
| − | NAME Apigenin 7-(3''-p-coumaroylglucoside) | + | NAME Apigenin 7-(3''-p-coumaroylglucoside) |
| − | CAS_RN - | + | CAS_RN - |
| − | FORMULA C30H26O12 | + | FORMULA C30H26O12 |
| − | EXACTMASS 578.1424262959999 | + | EXACTMASS 578.1424262959999 |
| − | AVERAGEMASS 578.5202400000001 | + | AVERAGEMASS 578.5202400000001 |
| − | SMILES c(O)(c5)ccc(c5)C(O4)=CC(c(c14)c(O)cc(OC(C2O)OC(CO)C(C2OC(=O)C=Cc(c3)ccc(O)c3)O)c1)=O | + | SMILES c(O)(c5)ccc(c5)C(O4)=CC(c(c14)c(O)cc(OC(C2O)OC(CO)C(C2OC(=O)C=Cc(c3)ccc(O)c3)O)c1)=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
-0.5988 2.0440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5988 1.2190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1157 0.8065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8302 1.2190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8302 2.0440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1157 2.4565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5446 0.8065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2591 1.2190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2591 2.0440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5446 2.4565 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9736 2.4565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6881 2.0440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4025 2.4565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4025 3.2815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6881 3.6940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9736 3.2815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5446 -0.0185 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1157 -0.0185 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.1488 3.7124 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3184 2.3910 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0279 2.4678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6153 1.7531 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8169 1.9623 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0234 1.7354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4360 2.4502 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2344 2.2411 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3914 2.8270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8712 3.1040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5111 1.9846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0639 2.0122 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0478 1.1783 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3250 1.2940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.1488 1.3023 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3328 0.4443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3379 -0.3810 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3417 -1.2386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0600 -1.6444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0678 -2.4694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3572 -2.8886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6389 -2.4828 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6312 -1.6578 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3649 -3.7124 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
7 17 2 0 0 0 0
3 18 1 0 0 0 0
14 19 1 0 0 0 0
20 1 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
26 27 1 0 0 0 0
27 28 1 0 0 0 0
21 29 1 0 0 0 0
22 30 1 0 0 0 0
24 20 1 0 0 0 0
32 33 2 0 0 0 0
32 34 1 0 0 0 0
34 35 2 0 0 0 0
35 36 1 0 0 0 0
36 37 2 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
39 40 1 0 0 0 0
40 41 2 0 0 0 0
41 36 1 0 0 0 0
39 42 1 0 0 0 0
31 23 1 0 0 0 0
32 30 1 0 0 0 0
S SKP 8
ID FL3FAAGS0065
KNApSAcK_ID C00013615
NAME Apigenin 7-(3''-p-coumaroylglucoside)
CAS_RN -
FORMULA C30H26O12
EXACTMASS 578.1424262959999
AVERAGEMASS 578.5202400000001
SMILES c(O)(c5)ccc(c5)C(O4)=CC(c(c14)c(O)cc(OC(C2O)OC(CO)C(C2OC(=O)C=Cc(c3)ccc(O)c3)O)c1)=O
M END