Mol:FL3FAAGS0045
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 53 58 0 0 0 0 0 0 0 0999 V2000 | + | 53 58 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.0196 0.5586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0196 0.5586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0196 -0.0144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0196 -0.0144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4767 -0.3009 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4767 -0.3009 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9729 -0.0144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9729 -0.0144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9729 0.5586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9729 0.5586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4767 0.8451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4767 0.8451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4691 -0.3009 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4691 -0.3009 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9653 -0.0144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9653 -0.0144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9653 0.5586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9653 0.5586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4691 0.8451 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4691 0.8451 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6579 -0.7056 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6579 -0.7056 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4614 0.8450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4614 0.8450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9671 0.5530 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9671 0.5530 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4728 0.8450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4728 0.8450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4728 1.4290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4728 1.4290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9671 1.7210 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9671 1.7210 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4614 1.4290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4614 1.4290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5156 0.8450 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5156 0.8450 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9784 1.7209 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9784 1.7209 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3992 -0.7410 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3992 -0.7410 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7992 0.5616 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -2.7992 0.5616 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -2.4115 0.0498 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.4115 0.0498 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.8531 0.2669 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.8531 0.2669 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.2408 0.1623 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.2408 0.1623 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.7059 0.6643 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7059 0.6643 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2762 0.4595 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -2.2762 0.4595 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -3.5916 0.2643 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5916 0.2643 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0159 0.0203 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0159 0.0203 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5332 -0.2702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5332 -0.2702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1941 0.8564 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1941 0.8564 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8370 1.4923 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8370 1.4923 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3749 1.9544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3749 1.9544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6420 2.4169 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6420 2.4169 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8424 1.9544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8424 1.9544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8045 2.3880 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8045 2.3880 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3341 2.6074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3341 2.6074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8583 2.2743 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8583 2.2743 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3318 2.5198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3318 2.5198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2812 3.0984 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2812 3.0984 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7570 3.4315 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7570 3.4315 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2834 3.1860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2834 3.1860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2446 3.3435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2446 3.3435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5332 -0.8806 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5332 -0.8806 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2291 -1.1847 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2291 -1.1847 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0866 -1.2001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0866 -1.2001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0866 -1.7421 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0866 -1.7421 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5253 -1.9954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5253 -1.9954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0097 -1.7157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0097 -1.7157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4941 -1.9954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4941 -1.9954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4941 -2.5547 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4941 -2.5547 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0097 -2.8344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0097 -2.8344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5253 -2.5547 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5253 -2.5547 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9784 -2.8343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9784 -2.8343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 15 19 1 0 0 0 0 | + | 15 19 1 0 0 0 0 |
| − | 20 3 1 0 0 0 0 | + | 20 3 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 18 24 1 0 0 0 0 | + | 18 24 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 2 0 0 0 0 | + | 32 33 2 0 0 0 0 |
| − | 32 34 1 0 0 0 0 | + | 32 34 1 0 0 0 0 |
| − | 34 35 2 0 0 0 0 | + | 34 35 2 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 2 0 0 0 0 | + | 40 41 2 0 0 0 0 |
| − | 41 36 1 0 0 0 0 | + | 41 36 1 0 0 0 0 |
| − | 39 42 1 0 0 0 0 | + | 39 42 1 0 0 0 0 |
| − | 29 43 1 0 0 0 0 | + | 29 43 1 0 0 0 0 |
| − | 43 44 2 0 0 0 0 | + | 43 44 2 0 0 0 0 |
| − | 43 45 1 0 0 0 0 | + | 43 45 1 0 0 0 0 |
| − | 45 46 2 0 0 0 0 | + | 45 46 2 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 2 0 0 0 0 | + | 47 48 2 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 49 50 2 0 0 0 0 | + | 49 50 2 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 51 52 2 0 0 0 0 | + | 51 52 2 0 0 0 0 |
| − | 52 47 1 0 0 0 0 | + | 52 47 1 0 0 0 0 |
| − | 50 53 1 0 0 0 0 | + | 50 53 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FAAGS0045 | + | ID FL3FAAGS0045 |
| − | KNApSAcK_ID C00004185 | + | KNApSAcK_ID C00004185 |
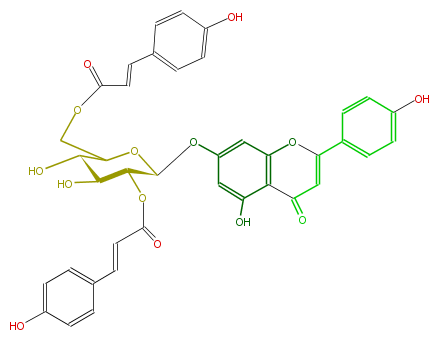
| − | NAME Apigenin 7-(2'',6''-di-p-coumarylglucoside) | + | NAME Apigenin 7-(2'',6''-di-p-coumarylglucoside) |
| − | CAS_RN 87562-08-1 | + | CAS_RN 87562-08-1 |
| − | FORMULA C39H32O14 | + | FORMULA C39H32O14 |
| − | EXACTMASS 724.179205732 | + | EXACTMASS 724.179205732 |
| − | AVERAGEMASS 724.6629800000001 | + | AVERAGEMASS 724.6629800000001 |
| − | SMILES C(c(c6)ccc(c6)O)=CC(=O)O[C@@H]([C@H]5O)[C@@H](OC([C@@H]5O)COC(=O)C=Cc(c4)ccc(c4)O)Oc(c3)cc(c(c(O)3)1)OC(c(c2)ccc(c2)O)=CC(=O)1 | + | SMILES C(c(c6)ccc(c6)O)=CC(=O)O[C@@H]([C@H]5O)[C@@H](OC([C@@H]5O)COC(=O)C=Cc(c4)ccc(c4)O)Oc(c3)cc(c(c(O)3)1)OC(c(c2)ccc(c2)O)=CC(=O)1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
53 58 0 0 0 0 0 0 0 0999 V2000
-0.0196 0.5586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0196 -0.0144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4767 -0.3009 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9729 -0.0144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9729 0.5586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4767 0.8451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4691 -0.3009 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9653 -0.0144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9653 0.5586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4691 0.8451 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6579 -0.7056 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4614 0.8450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9671 0.5530 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4728 0.8450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4728 1.4290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9671 1.7210 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4614 1.4290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5156 0.8450 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9784 1.7209 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3992 -0.7410 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7992 0.5616 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-2.4115 0.0498 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.8531 0.2669 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.2408 0.1623 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.7059 0.6643 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2762 0.4595 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.5916 0.2643 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0159 0.0203 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5332 -0.2702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1941 0.8564 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8370 1.4923 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3749 1.9544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6420 2.4169 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8424 1.9544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8045 2.3880 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3341 2.6074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8583 2.2743 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3318 2.5198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2812 3.0984 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7570 3.4315 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2834 3.1860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2446 3.3435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5332 -0.8806 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2291 -1.1847 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0866 -1.2001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0866 -1.7421 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5253 -1.9954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0097 -1.7157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4941 -1.9954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4941 -2.5547 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0097 -2.8344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5253 -2.5547 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9784 -2.8343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
15 19 1 0 0 0 0
20 3 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
26 30 1 0 0 0 0
18 24 1 0 0 0 0
30 31 1 0 0 0 0
31 32 1 0 0 0 0
32 33 2 0 0 0 0
32 34 1 0 0 0 0
34 35 2 0 0 0 0
35 36 1 0 0 0 0
36 37 2 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
39 40 1 0 0 0 0
40 41 2 0 0 0 0
41 36 1 0 0 0 0
39 42 1 0 0 0 0
29 43 1 0 0 0 0
43 44 2 0 0 0 0
43 45 1 0 0 0 0
45 46 2 0 0 0 0
46 47 1 0 0 0 0
47 48 2 0 0 0 0
48 49 1 0 0 0 0
49 50 2 0 0 0 0
50 51 1 0 0 0 0
51 52 2 0 0 0 0
52 47 1 0 0 0 0
50 53 1 0 0 0 0
S SKP 8
ID FL3FAAGS0045
KNApSAcK_ID C00004185
NAME Apigenin 7-(2'',6''-di-p-coumarylglucoside)
CAS_RN 87562-08-1
FORMULA C39H32O14
EXACTMASS 724.179205732
AVERAGEMASS 724.6629800000001
SMILES C(c(c6)ccc(c6)O)=CC(=O)O[C@@H]([C@H]5O)[C@@H](OC([C@@H]5O)COC(=O)C=Cc(c4)ccc(c4)O)Oc(c3)cc(c(c(O)3)1)OC(c(c2)ccc(c2)O)=CC(=O)1
M END