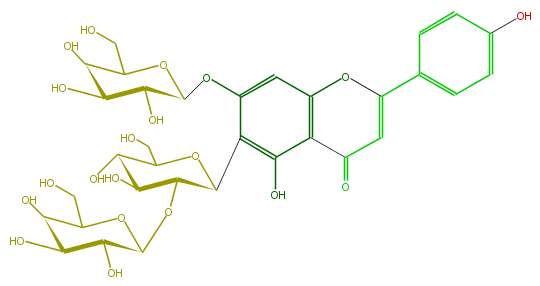
Mol:FL3FAADS0032
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 53 58 0 0 0 0 0 0 0 0999 V2000 | + | 53 58 0 0 0 0 0 0 0 0999 V2000 |
| − | 2.2316 0.9374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2316 0.9374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5171 1.3499 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5171 1.3499 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8026 0.9374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8026 0.9374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8026 0.1124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8026 0.1124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5171 -0.3001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5171 -0.3001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2316 0.1124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2316 0.1124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0881 1.3499 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0881 1.3499 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6263 0.9374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6263 0.9374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6263 0.1124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6263 0.1124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0881 -0.3001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0881 -0.3001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2974 1.3249 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2974 1.3249 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0881 -1.0395 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0881 -1.0395 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5171 -0.8721 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5171 -0.8721 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1018 -0.2336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1018 -0.2336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6892 -0.9483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6892 -0.9483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8908 -0.7391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8908 -0.7391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0973 -0.9660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0973 -0.9660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5099 -0.2512 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5099 -0.2512 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3083 -0.4603 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3083 -0.4603 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4713 -0.0794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4713 -0.0794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8169 0.1201 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8169 0.1201 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5850 -0.7168 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5850 -0.7168 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1378 -0.6893 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1378 -0.6893 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0623 -1.3791 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0623 -1.3791 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0133 1.3888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0133 1.3888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7278 0.9763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7278 0.9763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4422 1.3888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4422 1.3888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4422 2.2138 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4422 2.2138 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7278 2.6263 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7278 2.6263 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0133 2.2138 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0133 2.2138 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0575 2.5690 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0575 2.5690 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7632 1.6146 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7632 1.6146 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3506 0.8999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3506 0.8999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5523 1.1091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5523 1.1091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7588 0.8822 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7588 0.8822 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1714 1.5970 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1714 1.5970 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9698 1.3879 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9698 1.3879 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1381 2.0161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1381 2.0161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6181 2.2932 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6181 2.2932 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1109 1.9623 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1109 1.9623 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2160 1.1318 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2160 1.1318 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3826 0.4757 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3826 0.4757 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6047 -1.4874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6047 -1.4874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1921 -2.2021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1921 -2.2021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3938 -1.9929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3938 -1.9929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6003 -2.2198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6003 -2.2198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0129 -1.5050 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0129 -1.5050 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8113 -1.7141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8113 -1.7141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9796 -1.0859 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9796 -1.0859 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4596 -0.8088 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4596 -0.8088 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9524 -1.1397 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9524 -1.1397 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0575 -1.9702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0575 -1.9702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2241 -2.6263 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2241 -2.6263 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 3 7 1 0 0 0 0 | + | 3 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 4 1 0 0 0 0 | + | 10 4 1 0 0 0 0 |
| − | 8 11 1 0 0 0 0 | + | 8 11 1 0 0 0 0 |
| − | 10 12 1 0 0 0 0 | + | 10 12 1 0 0 0 0 |
| − | 5 13 2 0 0 0 0 | + | 5 13 2 0 0 0 0 |
| − | 14 15 1 1 0 0 0 | + | 14 15 1 1 0 0 0 |
| − | 15 16 1 1 0 0 0 | + | 15 16 1 1 0 0 0 |
| − | 17 16 1 1 0 0 0 | + | 17 16 1 1 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 19 1 0 0 0 0 | + | 18 19 1 0 0 0 0 |
| − | 19 14 1 0 0 0 0 | + | 19 14 1 0 0 0 0 |
| − | 19 20 1 0 0 0 0 | + | 19 20 1 0 0 0 0 |
| − | 20 21 1 0 0 0 0 | + | 20 21 1 0 0 0 0 |
| − | 14 22 1 0 0 0 0 | + | 14 22 1 0 0 0 0 |
| − | 15 23 1 0 0 0 0 | + | 15 23 1 0 0 0 0 |
| − | 16 24 1 0 0 0 0 | + | 16 24 1 0 0 0 0 |
| − | 9 17 1 0 0 0 0 | + | 9 17 1 0 0 0 0 |
| − | 1 25 1 0 0 0 0 | + | 1 25 1 0 0 0 0 |
| − | 25 26 2 0 0 0 0 | + | 25 26 2 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 2 0 0 0 0 | + | 27 28 2 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 30 2 0 0 0 0 | + | 29 30 2 0 0 0 0 |
| − | 30 25 1 0 0 0 0 | + | 30 25 1 0 0 0 0 |
| − | 28 31 1 0 0 0 0 | + | 28 31 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 32 40 1 0 0 0 0 | + | 32 40 1 0 0 0 0 |
| − | 33 41 1 0 0 0 0 | + | 33 41 1 0 0 0 0 |
| − | 34 42 1 0 0 0 0 | + | 34 42 1 0 0 0 0 |
| − | 43 44 1 1 0 0 0 | + | 43 44 1 1 0 0 0 |
| − | 44 45 1 1 0 0 0 | + | 44 45 1 1 0 0 0 |
| − | 46 45 1 1 0 0 0 | + | 46 45 1 1 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 48 43 1 0 0 0 0 | + | 48 43 1 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 49 50 1 0 0 0 0 | + | 49 50 1 0 0 0 0 |
| − | 43 51 1 0 0 0 0 | + | 43 51 1 0 0 0 0 |
| − | 44 52 1 0 0 0 0 | + | 44 52 1 0 0 0 0 |
| − | 45 53 1 0 0 0 0 | + | 45 53 1 0 0 0 0 |
| − | 46 24 1 0 0 0 0 | + | 46 24 1 0 0 0 0 |
| − | 35 11 1 0 0 0 0 | + | 35 11 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FAADS0032 | + | ID FL3FAADS0032 |
| − | KNApSAcK_ID C00014032 | + | KNApSAcK_ID C00014032 |
| − | NAME Isovitexin 7,2''-Di-O-galactoside | + | NAME Isovitexin 7,2''-Di-O-galactoside |
| − | CAS_RN - | + | CAS_RN - |
| − | FORMULA C33H40O20 | + | FORMULA C33H40O20 |
| − | EXACTMASS 756.21129372 | + | EXACTMASS 756.21129372 |
| − | AVERAGEMASS 756.6587 | + | AVERAGEMASS 756.6587 |
| − | SMILES C(=C5)(c(c6)ccc(O)c6)Oc(c(C5=O)1)cc(OC(O4)C(C(O)C(O)C4CO)O)c(C(C2OC(C3O)OC(C(C3O)O)CO)OC(CO)C(C(O)2)O)c1O | + | SMILES C(=C5)(c(c6)ccc(O)c6)Oc(c(C5=O)1)cc(OC(O4)C(C(O)C(O)C4CO)O)c(C(C2OC(C3O)OC(C(C3O)O)CO)OC(CO)C(C(O)2)O)c1O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
53 58 0 0 0 0 0 0 0 0999 V2000
2.2316 0.9374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5171 1.3499 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8026 0.9374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8026 0.1124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5171 -0.3001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2316 0.1124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0881 1.3499 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6263 0.9374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6263 0.1124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0881 -0.3001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2974 1.3249 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0881 -1.0395 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5171 -0.8721 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1018 -0.2336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6892 -0.9483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8908 -0.7391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0973 -0.9660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5099 -0.2512 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3083 -0.4603 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4713 -0.0794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8169 0.1201 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5850 -0.7168 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1378 -0.6893 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0623 -1.3791 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0133 1.3888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7278 0.9763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4422 1.3888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4422 2.2138 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7278 2.6263 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0133 2.2138 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.0575 2.5690 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7632 1.6146 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3506 0.8999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5523 1.1091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7588 0.8822 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1714 1.5970 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9698 1.3879 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1381 2.0161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6181 2.2932 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1109 1.9623 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2160 1.1318 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3826 0.4757 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6047 -1.4874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1921 -2.2021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3938 -1.9929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6003 -2.2198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0129 -1.5050 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8113 -1.7141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9796 -1.0859 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4596 -0.8088 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9524 -1.1397 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.0575 -1.9702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2241 -2.6263 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
3 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 4 1 0 0 0 0
8 11 1 0 0 0 0
10 12 1 0 0 0 0
5 13 2 0 0 0 0
14 15 1 1 0 0 0
15 16 1 1 0 0 0
17 16 1 1 0 0 0
17 18 1 0 0 0 0
18 19 1 0 0 0 0
19 14 1 0 0 0 0
19 20 1 0 0 0 0
20 21 1 0 0 0 0
14 22 1 0 0 0 0
15 23 1 0 0 0 0
16 24 1 0 0 0 0
9 17 1 0 0 0 0
1 25 1 0 0 0 0
25 26 2 0 0 0 0
26 27 1 0 0 0 0
27 28 2 0 0 0 0
28 29 1 0 0 0 0
29 30 2 0 0 0 0
30 25 1 0 0 0 0
28 31 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
37 38 1 0 0 0 0
38 39 1 0 0 0 0
32 40 1 0 0 0 0
33 41 1 0 0 0 0
34 42 1 0 0 0 0
43 44 1 1 0 0 0
44 45 1 1 0 0 0
46 45 1 1 0 0 0
46 47 1 0 0 0 0
47 48 1 0 0 0 0
48 43 1 0 0 0 0
48 49 1 0 0 0 0
49 50 1 0 0 0 0
43 51 1 0 0 0 0
44 52 1 0 0 0 0
45 53 1 0 0 0 0
46 24 1 0 0 0 0
35 11 1 0 0 0 0
S SKP 8
ID FL3FAADS0032
KNApSAcK_ID C00014032
NAME Isovitexin 7,2''-Di-O-galactoside
CAS_RN -
FORMULA C33H40O20
EXACTMASS 756.21129372
AVERAGEMASS 756.6587
SMILES C(=C5)(c(c6)ccc(O)c6)Oc(c(C5=O)1)cc(OC(O4)C(C(O)C(O)C4CO)O)c(C(C2OC(C3O)OC(C(C3O)O)CO)OC(CO)C(C(O)2)O)c1O
M END