Mol:FL2FABGM0004
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 56 61 0 0 0 0 0 0 0 0999 V2000 | + | 56 61 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.6415 0.0838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6415 0.0838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3541 0.4994 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3541 0.4994 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6447 -0.7412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6447 -0.7412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3614 -1.1506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3614 -1.1506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0741 -0.7349 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0741 -0.7349 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0704 0.0901 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0704 0.0901 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7904 -1.1442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7904 -1.1442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5030 -0.7286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5030 -0.7286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4993 0.0964 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4993 0.0964 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7830 0.5057 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7830 0.5057 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1531 0.4772 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1531 0.4772 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8692 0.0675 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8692 0.0675 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.5820 0.4828 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.5820 0.4828 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.5789 1.3078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.5789 1.3078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8628 1.7175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8628 1.7175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1499 1.3022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1499 1.3022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7914 -1.7875 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7914 -1.7875 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0160 0.4449 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0160 0.4449 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3614 -1.8587 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3614 -1.8587 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0525 -1.1437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0525 -1.1437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3541 1.2461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3541 1.2461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8703 0.6133 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8703 0.6133 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4576 -0.1014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4576 -0.1014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6593 0.1078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6593 0.1078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8658 -0.1191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8658 -0.1191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2784 0.5957 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2784 0.5957 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0768 0.3866 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0768 0.3866 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2337 0.9724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2337 0.9724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3535 0.1301 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3535 0.1301 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9063 0.1577 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9063 0.1577 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8308 -0.5322 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8308 -0.5322 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.1658 1.6466 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.1658 1.6466 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.7692 1.2982 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.7692 1.2982 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7287 1.5299 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7287 1.5299 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2963 2.2326 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2963 2.2326 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6887 2.9583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6887 2.9583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.5134 2.9814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.5134 2.9814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.9457 2.2787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.9457 2.2787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.5533 1.5530 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.5533 1.5530 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.9851 0.8513 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.9851 0.8513 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.7692 2.3018 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.7692 2.3018 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.9052 3.7061 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.9052 3.7061 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4728 2.2096 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4728 2.2096 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0410 2.9113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0410 2.9113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0809 1.4848 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0809 1.4848 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0001 -1.6538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0001 -1.6538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7094 -1.2325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7094 -1.2325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4290 -1.6360 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4290 -1.6360 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4393 -2.4609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4393 -2.4609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7300 -2.8823 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7300 -2.8823 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0104 -2.4788 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0104 -2.4788 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3021 -2.8996 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3021 -2.8996 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7403 -3.7061 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7403 -3.7061 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1579 -2.8639 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1579 -2.8639 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6991 -0.4086 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6991 -0.4086 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4074 0.0122 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4074 0.0122 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 1 3 1 0 0 0 0 | + | 1 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 2 1 0 0 0 0 | + | 6 2 1 0 0 0 0 |
| − | 5 7 1 0 0 0 0 | + | 5 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 6 1 0 0 0 0 | + | 10 6 1 0 0 0 0 |
| − | 9 11 1 6 0 0 0 | + | 9 11 1 6 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 7 17 2 0 0 0 0 | + | 7 17 2 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 4 19 1 0 0 0 0 | + | 4 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 2 21 1 0 0 0 0 | + | 2 21 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 22 29 1 0 0 0 0 | + | 22 29 1 0 0 0 0 |
| − | 23 30 1 0 0 0 0 | + | 23 30 1 0 0 0 0 |
| − | 24 31 1 0 0 0 0 | + | 24 31 1 0 0 0 0 |
| − | 14 32 1 0 0 0 0 | + | 14 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 25 18 1 0 0 0 0 | + | 25 18 1 0 0 0 0 |
| − | 34 35 2 0 0 0 0 | + | 34 35 2 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 39 34 1 0 0 0 0 | + | 39 34 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 38 41 1 0 0 0 0 | + | 38 41 1 0 0 0 0 |
| − | 37 42 1 0 0 0 0 | + | 37 42 1 0 0 0 0 |
| − | 35 43 1 0 0 0 0 | + | 35 43 1 0 0 0 0 |
| − | 43 44 2 0 0 0 0 | + | 43 44 2 0 0 0 0 |
| − | 43 45 1 0 0 0 0 | + | 43 45 1 0 0 0 0 |
| − | 45 28 1 0 0 0 0 | + | 45 28 1 0 0 0 0 |
| − | 46 47 2 0 0 0 0 | + | 46 47 2 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 48 49 2 0 0 0 0 | + | 48 49 2 0 0 0 0 |
| − | 49 50 1 0 0 0 0 | + | 49 50 1 0 0 0 0 |
| − | 50 51 2 0 0 0 0 | + | 50 51 2 0 0 0 0 |
| − | 51 46 1 0 0 0 0 | + | 51 46 1 0 0 0 0 |
| − | 51 52 1 0 0 0 0 | + | 51 52 1 0 0 0 0 |
| − | 50 53 1 0 0 0 0 | + | 50 53 1 0 0 0 0 |
| − | 49 54 1 0 0 0 0 | + | 49 54 1 0 0 0 0 |
| − | 47 55 1 0 0 0 0 | + | 47 55 1 0 0 0 0 |
| − | 55 56 2 0 0 0 0 | + | 55 56 2 0 0 0 0 |
| − | 55 29 1 0 0 0 0 | + | 55 29 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL2FABGM0004 | + | ID FL2FABGM0004 |
| − | KNApSAcK_ID C00014357 | + | KNApSAcK_ID C00014357 |
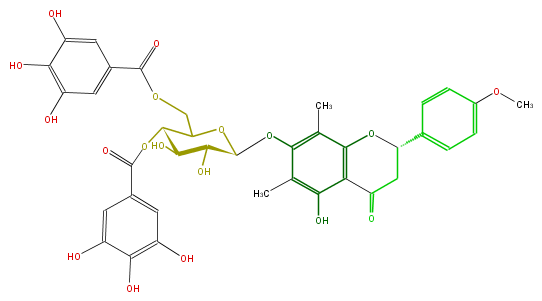
| − | NAME 5,7-Dihydroxy-4'-methoxy-6,8-di-C-methylflavanone 7-(4,6-digalloylglucoside);Matteucinol-7-O-[4'',6''-di-O-galloyl]-beta-D-glucopyranoside | + | NAME 5,7-Dihydroxy-4'-methoxy-6,8-di-C-methylflavanone 7-(4,6-digalloylglucoside);Matteucinol-7-O-[4'',6''-di-O-galloyl]-beta-D-glucopyranoside |
| − | CAS_RN 374907-39-8 | + | CAS_RN 374907-39-8 |
| − | FORMULA C38H36O18 | + | FORMULA C38H36O18 |
| − | EXACTMASS 780.190164348 | + | EXACTMASS 780.190164348 |
| − | AVERAGEMASS 780.68164 | + | AVERAGEMASS 780.68164 |
| − | SMILES c(c(O)6)c(cc(O)c6O)C(=O)OC(C4COC(c(c5)cc(O)c(c(O)5)O)=O)C(C(O)C(O4)Oc(c3C)c(C)c(c(c(O)3)2)OC(CC2=O)c(c1)ccc(OC)c1)O | + | SMILES c(c(O)6)c(cc(O)c6O)C(=O)OC(C4COC(c(c5)cc(O)c(c(O)5)O)=O)C(C(O)C(O4)Oc(c3C)c(C)c(c(c(O)3)2)OC(CC2=O)c(c1)ccc(OC)c1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
56 61 0 0 0 0 0 0 0 0999 V2000
0.6415 0.0838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3541 0.4994 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6447 -0.7412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3614 -1.1506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0741 -0.7349 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0704 0.0901 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7904 -1.1442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5030 -0.7286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4993 0.0964 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7830 0.5057 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1531 0.4772 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8692 0.0675 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.5820 0.4828 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.5789 1.3078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8628 1.7175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1499 1.3022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7914 -1.7875 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0160 0.4449 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3614 -1.8587 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0525 -1.1437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3541 1.2461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8703 0.6133 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4576 -0.1014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6593 0.1078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8658 -0.1191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2784 0.5957 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0768 0.3866 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2337 0.9724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3535 0.1301 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9063 0.1577 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8308 -0.5322 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.1658 1.6466 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.7692 1.2982 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7287 1.5299 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2963 2.2326 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6887 2.9583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.5134 2.9814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.9457 2.2787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.5533 1.5530 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.9851 0.8513 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-6.7692 2.3018 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.9052 3.7061 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4728 2.2096 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0410 2.9113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0809 1.4848 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0001 -1.6538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7094 -1.2325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4290 -1.6360 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4393 -2.4609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7300 -2.8823 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0104 -2.4788 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3021 -2.8996 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7403 -3.7061 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.1579 -2.8639 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6991 -0.4086 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4074 0.0122 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
1 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 2 1 0 0 0 0
5 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 1 0 0 0 0
10 6 1 0 0 0 0
9 11 1 6 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
7 17 2 0 0 0 0
1 18 1 0 0 0 0
4 19 1 0 0 0 0
3 20 1 0 0 0 0
2 21 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
27 28 1 0 0 0 0
22 29 1 0 0 0 0
23 30 1 0 0 0 0
24 31 1 0 0 0 0
14 32 1 0 0 0 0
32 33 1 0 0 0 0
25 18 1 0 0 0 0
34 35 2 0 0 0 0
35 36 1 0 0 0 0
36 37 2 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
39 34 1 0 0 0 0
39 40 1 0 0 0 0
38 41 1 0 0 0 0
37 42 1 0 0 0 0
35 43 1 0 0 0 0
43 44 2 0 0 0 0
43 45 1 0 0 0 0
45 28 1 0 0 0 0
46 47 2 0 0 0 0
47 48 1 0 0 0 0
48 49 2 0 0 0 0
49 50 1 0 0 0 0
50 51 2 0 0 0 0
51 46 1 0 0 0 0
51 52 1 0 0 0 0
50 53 1 0 0 0 0
49 54 1 0 0 0 0
47 55 1 0 0 0 0
55 56 2 0 0 0 0
55 29 1 0 0 0 0
S SKP 8
ID FL2FABGM0004
KNApSAcK_ID C00014357
NAME 5,7-Dihydroxy-4'-methoxy-6,8-di-C-methylflavanone 7-(4,6-digalloylglucoside);Matteucinol-7-O-[4'',6''-di-O-galloyl]-beta-D-glucopyranoside
CAS_RN 374907-39-8
FORMULA C38H36O18
EXACTMASS 780.190164348
AVERAGEMASS 780.68164
SMILES c(c(O)6)c(cc(O)c6O)C(=O)OC(C4COC(c(c5)cc(O)c(c(O)5)O)=O)C(C(O)C(O4)Oc(c3C)c(C)c(c(c(O)3)2)OC(CC2=O)c(c1)ccc(OC)c1)O
M END