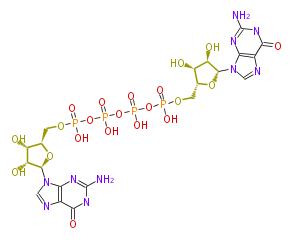
Mol:BMCCPUGU0018
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 55 60 0 0 1 0 0 0 0 0999 V2000 | + | 55 60 0 0 1 0 0 0 0 0999 V2000 |
| − | 3.7321 1.6347 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7321 1.6347 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5981 1.1347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5981 1.1347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5981 0.1347 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5981 0.1347 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7321 -0.3653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7321 -0.3653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8660 0.1347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8660 0.1347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8660 1.1347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8660 1.1347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1229 -0.5344 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1229 -0.5344 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5296 -1.4480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5296 -1.4480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5241 -1.3434 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5241 -1.3434 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4641 1.6347 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4641 1.6347 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0000 1.6347 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0000 1.6347 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1933 -2.0866 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 4.1933 -2.0866 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.9854 -3.0647 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.9854 -3.0647 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 4.8514 -3.5647 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 4.8514 -3.5647 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 5.5945 -2.8956 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 5.5945 -2.8956 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 6.5727 -3.1035 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.5727 -3.1035 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0718 -3.4715 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0718 -3.4715 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9559 -4.5593 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9559 -4.5593 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1878 -1.9821 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1878 -1.9821 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.2418 -2.3604 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.2418 -2.3604 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 19.9535 3.2721 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 19.9535 3.2721 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 19.0025 3.5811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 19.0025 3.5811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 18.2593 2.9120 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 18.2593 2.9120 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 18.4672 1.9338 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 18.4672 1.9338 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 19.4183 1.6248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 19.4183 1.6248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 20.1614 2.2939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 20.1614 2.2939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 19.4183 0.6248 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 19.4183 0.6248 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 18.4672 0.3158 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 18.4672 0.3158 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 17.8795 1.1248 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 17.8795 1.1248 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 18.7946 4.5593 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 18.7946 4.5593 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 21.1614 2.2939 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 21.1614 2.2939 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 16.8795 1.1248 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 16.8795 1.1248 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 16.1363 1.7939 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 16.1363 1.7939 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 15.2703 1.2939 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 15.2703 1.2939 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 15.4782 0.3158 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 15.4782 0.3158 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 14.8091 -0.4273 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 14.8091 -0.4273 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 16.2408 2.7885 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 16.2408 2.7885 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 14.3567 1.7007 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 14.3567 1.7007 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 16.4727 0.2113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 16.4727 0.2113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 13.8309 -0.2194 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 13.8309 -0.2194 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 13.1618 -0.9626 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 | + | 13.1618 -0.9626 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 13.9049 -1.6317 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 13.9049 -1.6317 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 12.4186 -0.2935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 12.4186 -0.2935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 12.4927 -1.7057 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 12.4927 -1.7057 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 11.5145 -1.4978 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 | + | 11.5145 -1.4978 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 11.3066 -2.4760 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 11.3066 -2.4760 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 11.7224 -0.5197 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 11.7224 -0.5197 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 10.5364 -1.2899 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 10.5364 -1.2899 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.8672 -2.0330 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.8672 -2.0330 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 10.6104 -2.7022 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 10.6104 -2.7022 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.1241 -1.3639 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.1241 -1.3639 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.1981 -2.7762 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.1981 -2.7762 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 8.2200 -2.5683 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 | + | 8.2200 -2.5683 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 8.0120 -3.5464 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 8.0120 -3.5464 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 8.4279 -1.5901 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 8.4279 -1.5901 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 3 2 2 0 0 0 0 | + | 3 2 2 0 0 0 0 |
| − | 2 1 1 0 0 0 0 | + | 2 1 1 0 0 0 0 |
| − | 1 6 1 0 0 0 0 | + | 1 6 1 0 0 0 0 |
| − | 6 5 1 0 0 0 0 | + | 6 5 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 7 1 0 0 0 0 | + | 5 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 4 1 0 0 0 0 | + | 9 4 1 0 0 0 0 |
| − | 6 11 2 0 0 0 0 | + | 6 11 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 19 1 6 0 0 0 | + | 15 19 1 6 0 0 0 |
| − | 12 19 1 6 0 0 0 | + | 12 19 1 6 0 0 0 |
| − | 13 17 1 1 0 0 0 | + | 13 17 1 1 0 0 0 |
| − | 14 18 1 1 0 0 0 | + | 14 18 1 1 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 20 1 0 0 0 0 | + | 16 20 1 0 0 0 0 |
| − | 12 9 1 0 0 0 0 | + | 12 9 1 0 0 0 0 |
| − | 24 23 1 0 0 0 0 | + | 24 23 1 0 0 0 0 |
| − | 23 22 2 0 0 0 0 | + | 23 22 2 0 0 0 0 |
| − | 22 21 1 0 0 0 0 | + | 22 21 1 0 0 0 0 |
| − | 21 26 1 0 0 0 0 | + | 21 26 1 0 0 0 0 |
| − | 26 25 1 0 0 0 0 | + | 26 25 1 0 0 0 0 |
| − | 24 25 2 0 0 0 0 | + | 24 25 2 0 0 0 0 |
| − | 25 27 1 0 0 0 0 | + | 25 27 1 0 0 0 0 |
| − | 27 28 2 0 0 0 0 | + | 27 28 2 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 24 1 0 0 0 0 | + | 29 24 1 0 0 0 0 |
| − | 26 31 2 0 0 0 0 | + | 26 31 2 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 39 1 6 0 0 0 | + | 35 39 1 6 0 0 0 |
| − | 32 39 1 6 0 0 0 | + | 32 39 1 6 0 0 0 |
| − | 33 37 1 1 0 0 0 | + | 33 37 1 1 0 0 0 |
| − | 34 38 1 1 0 0 0 | + | 34 38 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 32 29 1 0 0 0 0 | + | 32 29 1 0 0 0 0 |
| − | 20 53 1 0 0 0 0 | + | 20 53 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 53 54 1 0 0 0 0 | + | 53 54 1 0 0 0 0 |
| − | 53 55 2 0 0 0 0 | + | 53 55 2 0 0 0 0 |
| − | 52 49 1 0 0 0 0 | + | 52 49 1 0 0 0 0 |
| − | 49 48 1 0 0 0 0 | + | 49 48 1 0 0 0 0 |
| − | 49 50 1 0 0 0 0 | + | 49 50 1 0 0 0 0 |
| − | 49 51 2 0 0 0 0 | + | 49 51 2 0 0 0 0 |
| − | 48 45 1 0 0 0 0 | + | 48 45 1 0 0 0 0 |
| − | 45 44 1 0 0 0 0 | + | 45 44 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 45 47 2 0 0 0 0 | + | 45 47 2 0 0 0 0 |
| − | 44 41 1 0 0 0 0 | + | 44 41 1 0 0 0 0 |
| − | 41 40 1 0 0 0 0 | + | 41 40 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 41 43 2 0 0 0 0 | + | 41 43 2 0 0 0 0 |
| − | 2 10 1 0 0 0 0 | + | 2 10 1 0 0 0 0 |
| − | 22 30 1 0 0 0 0 | + | 22 30 1 0 0 0 0 |
| − | S SKP 7 | + | S SKP 7 |
| − | ID BMCCPUGU0018 | + | ID BMCCPUGU0018 |
| − | NAME P1,P4-Bis(5'-guanosyl) tetraphosphate | + | NAME P1,P4-Bis(5'-guanosyl) tetraphosphate |
| − | FORMULA C20H28N10O21P4 | + | FORMULA C20H28N10O21P4 |
| − | EXACTMASS 868.038 | + | EXACTMASS 868.038 |
| − | AVERAGEMASS 868.3861 | + | AVERAGEMASS 868.3861 |
| − | SMILES O([C@@H]1COP(O)(=O)OP(OP(O)(=O)OP(O)(=O)OC[C@@H](O4)[C@H]([C@@H](O)[C@@H]4n(c6)c(c(n6)5)N=C(NC(=O)5)N)O)(O)=O)[C@@H](n(c3)c(c(n3)2)N=C(NC(=O)2)N)[C@@H]([C@@H]1O)O | + | SMILES O([C@@H]1COP(O)(=O)OP(OP(O)(=O)OP(O)(=O)OC[C@@H](O4)[C@H]([C@@H](O)[C@@H]4n(c6)c(c(n6)5)N=C(NC(=O)5)N)O)(O)=O)[C@@H](n(c3)c(c(n3)2)N=C(NC(=O)2)N)[C@@H]([C@@H]1O)O |
| − | KEGG http://www.genome.jp/dbget-bin/www_bget?compound+C01261 | + | KEGG http://www.genome.jp/dbget-bin/www_bget?compound+C01261 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
55 60 0 0 1 0 0 0 0 0999 V2000
3.7321 1.6347 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
4.5981 1.1347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5981 0.1347 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
3.7321 -0.3653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8660 0.1347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8660 1.1347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1229 -0.5344 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
2.5296 -1.4480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5241 -1.3434 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
5.4641 1.6347 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
2.0000 1.6347 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1933 -2.0866 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.9854 -3.0647 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
4.8514 -3.5647 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
5.5945 -2.8956 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
6.5727 -3.1035 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0718 -3.4715 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9559 -4.5593 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.1878 -1.9821 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
7.2418 -2.3604 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
19.9535 3.2721 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
19.0025 3.5811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
18.2593 2.9120 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
18.4672 1.9338 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
19.4183 1.6248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
20.1614 2.2939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
19.4183 0.6248 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
18.4672 0.3158 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
17.8795 1.1248 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
18.7946 4.5593 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
21.1614 2.2939 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
16.8795 1.1248 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
16.1363 1.7939 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
15.2703 1.2939 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
15.4782 0.3158 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
14.8091 -0.4273 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
16.2408 2.7885 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
14.3567 1.7007 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
16.4727 0.2113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
13.8309 -0.2194 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
13.1618 -0.9626 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0
13.9049 -1.6317 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
12.4186 -0.2935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
12.4927 -1.7057 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
11.5145 -1.4978 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0
11.3066 -2.4760 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
11.7224 -0.5197 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
10.5364 -1.2899 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
9.8672 -2.0330 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0
10.6104 -2.7022 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
9.1241 -1.3639 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
9.1981 -2.7762 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
8.2200 -2.5683 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0
8.0120 -3.5464 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
8.4279 -1.5901 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4 3 1 0 0 0 0
3 2 2 0 0 0 0
2 1 1 0 0 0 0
1 6 1 0 0 0 0
6 5 1 0 0 0 0
4 5 2 0 0 0 0
5 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 4 1 0 0 0 0
6 11 2 0 0 0 0
12 13 1 0 0 0 0
13 14 1 0 0 0 0
14 15 1 0 0 0 0
15 19 1 6 0 0 0
12 19 1 6 0 0 0
13 17 1 1 0 0 0
14 18 1 1 0 0 0
15 16 1 0 0 0 0
16 20 1 0 0 0 0
12 9 1 0 0 0 0
24 23 1 0 0 0 0
23 22 2 0 0 0 0
22 21 1 0 0 0 0
21 26 1 0 0 0 0
26 25 1 0 0 0 0
24 25 2 0 0 0 0
25 27 1 0 0 0 0
27 28 2 0 0 0 0
28 29 1 0 0 0 0
29 24 1 0 0 0 0
26 31 2 0 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 39 1 6 0 0 0
32 39 1 6 0 0 0
33 37 1 1 0 0 0
34 38 1 1 0 0 0
35 36 1 0 0 0 0
36 40 1 0 0 0 0
32 29 1 0 0 0 0
20 53 1 0 0 0 0
52 53 1 0 0 0 0
53 54 1 0 0 0 0
53 55 2 0 0 0 0
52 49 1 0 0 0 0
49 48 1 0 0 0 0
49 50 1 0 0 0 0
49 51 2 0 0 0 0
48 45 1 0 0 0 0
45 44 1 0 0 0 0
45 46 1 0 0 0 0
45 47 2 0 0 0 0
44 41 1 0 0 0 0
41 40 1 0 0 0 0
41 42 1 0 0 0 0
41 43 2 0 0 0 0
2 10 1 0 0 0 0
22 30 1 0 0 0 0
S SKP 7
ID BMCCPUGU0018
NAME P1,P4-Bis(5'-guanosyl) tetraphosphate
FORMULA C20H28N10O21P4
EXACTMASS 868.038
AVERAGEMASS 868.3861
SMILES O([C@@H]1COP(O)(=O)OP(OP(O)(=O)OP(O)(=O)OC[C@@H](O4)[C@H]([C@@H](O)[C@@H]4n(c6)c(c(n6)5)N=C(NC(=O)5)N)O)(O)=O)[C@@H](n(c3)c(c(n3)2)N=C(NC(=O)2)N)[C@@H]([C@@H]1O)O
KEGG http://www.genome.jp/dbget-bin/www_bget?compound+C01261
M END