Mol:BMCCPUAP0037
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 53 58 0 0 1 0 0 0 0 0999 V2000 | + | 53 58 0 0 1 0 0 0 0 0999 V2000 |
| − | 3.7321 2.1238 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7321 2.1238 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5981 1.6238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5981 1.6238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5981 0.6238 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5981 0.6238 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7321 0.1238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7321 0.1238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8660 0.6238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8660 0.6238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8660 1.6238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8660 1.6238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1229 -0.0454 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1229 -0.0454 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5296 -0.9589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5296 -0.9589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5241 -0.8544 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5241 -0.8544 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0000 2.1238 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0000 2.1238 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1933 -1.5975 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 4.1933 -1.5975 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.9854 -2.5757 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.9854 -2.5757 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 4.8514 -3.0757 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 4.8514 -3.0757 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 5.5945 -2.4065 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 5.5945 -2.4065 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 6.5727 -2.6144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.5727 -2.6144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0718 -2.9824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0718 -2.9824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9559 -4.0702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9559 -4.0702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1878 -1.4930 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1878 -1.4930 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.2418 -1.8713 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.2418 -1.8713 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 19.9535 3.7612 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 19.9535 3.7612 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 19.0025 4.0702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 19.0025 4.0702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 18.2593 3.4011 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 18.2593 3.4011 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 18.4672 2.4229 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 18.4672 2.4229 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 19.4183 2.1139 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 19.4183 2.1139 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 20.1614 2.7830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 20.1614 2.7830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 19.4183 1.1139 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 19.4183 1.1139 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 18.4672 0.8049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 18.4672 0.8049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 17.8795 1.6139 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 17.8795 1.6139 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 21.1614 2.7830 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 21.1614 2.7830 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 16.8795 1.6139 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 16.8795 1.6139 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 16.1363 2.2830 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 16.1363 2.2830 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 15.2703 1.7830 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 15.2703 1.7830 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 15.4782 0.8049 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 15.4782 0.8049 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 14.8091 0.0617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 14.8091 0.0617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 16.2408 3.2775 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 16.2408 3.2775 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 14.3567 2.1898 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 14.3567 2.1898 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 16.4727 0.7003 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 16.4727 0.7003 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 13.8309 0.2696 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 13.8309 0.2696 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 13.1618 -0.4735 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 | + | 13.1618 -0.4735 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 13.9049 -1.1426 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 13.9049 -1.1426 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 12.4186 0.1956 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 12.4186 0.1956 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 12.4927 -1.2167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 12.4927 -1.2167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 11.5145 -1.0087 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 | + | 11.5145 -1.0087 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 11.3066 -1.9869 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 11.3066 -1.9869 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 11.7224 -0.0306 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 11.7224 -0.0306 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 10.5364 -0.8008 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 10.5364 -0.8008 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.8672 -1.5440 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.8672 -1.5440 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 10.6104 -2.2131 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 10.6104 -2.2131 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.1241 -0.8748 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.1241 -0.8748 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.1981 -2.2871 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.1981 -2.2871 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 8.2200 -2.0792 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 | + | 8.2200 -2.0792 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 8.0120 -3.0574 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 8.0120 -3.0574 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 8.4279 -1.1011 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 8.4279 -1.1011 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 3 2 2 0 0 0 0 | + | 3 2 2 0 0 0 0 |
| − | 2 1 1 0 0 0 0 | + | 2 1 1 0 0 0 0 |
| − | 1 6 2 0 0 0 0 | + | 1 6 2 0 0 0 0 |
| − | 6 5 1 0 0 0 0 | + | 6 5 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 7 1 0 0 0 0 | + | 5 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 4 1 0 0 0 0 | + | 9 4 1 0 0 0 0 |
| − | 6 10 1 0 0 0 0 | + | 6 10 1 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 18 1 6 0 0 0 | + | 14 18 1 6 0 0 0 |
| − | 11 18 1 6 0 0 0 | + | 11 18 1 6 0 0 0 |
| − | 12 16 1 1 0 0 0 | + | 12 16 1 1 0 0 0 |
| − | 13 17 1 1 0 0 0 | + | 13 17 1 1 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 19 1 0 0 0 0 | + | 15 19 1 0 0 0 0 |
| − | 11 9 1 0 0 0 0 | + | 11 9 1 0 0 0 0 |
| − | 23 22 1 0 0 0 0 | + | 23 22 1 0 0 0 0 |
| − | 22 21 2 0 0 0 0 | + | 22 21 2 0 0 0 0 |
| − | 21 20 1 0 0 0 0 | + | 21 20 1 0 0 0 0 |
| − | 20 25 2 0 0 0 0 | + | 20 25 2 0 0 0 0 |
| − | 25 24 1 0 0 0 0 | + | 25 24 1 0 0 0 0 |
| − | 23 24 2 0 0 0 0 | + | 23 24 2 0 0 0 0 |
| − | 24 26 1 0 0 0 0 | + | 24 26 1 0 0 0 0 |
| − | 26 27 2 0 0 0 0 | + | 26 27 2 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 37 1 6 0 0 0 | + | 33 37 1 6 0 0 0 |
| − | 30 37 1 6 0 0 0 | + | 30 37 1 6 0 0 0 |
| − | 31 35 1 1 0 0 0 | + | 31 35 1 1 0 0 0 |
| − | 32 36 1 1 0 0 0 | + | 32 36 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 38 1 0 0 0 0 | + | 34 38 1 0 0 0 0 |
| − | 30 28 1 0 0 0 0 | + | 30 28 1 0 0 0 0 |
| − | 19 51 1 0 0 0 0 | + | 19 51 1 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 51 52 1 0 0 0 0 | + | 51 52 1 0 0 0 0 |
| − | 51 53 2 0 0 0 0 | + | 51 53 2 0 0 0 0 |
| − | 50 47 1 0 0 0 0 | + | 50 47 1 0 0 0 0 |
| − | 47 46 1 0 0 0 0 | + | 47 46 1 0 0 0 0 |
| − | 47 49 1 0 0 0 0 | + | 47 49 1 0 0 0 0 |
| − | 47 48 2 0 0 0 0 | + | 47 48 2 0 0 0 0 |
| − | 46 43 1 0 0 0 0 | + | 46 43 1 0 0 0 0 |
| − | 43 42 1 0 0 0 0 | + | 43 42 1 0 0 0 0 |
| − | 43 45 1 0 0 0 0 | + | 43 45 1 0 0 0 0 |
| − | 43 44 2 0 0 0 0 | + | 43 44 2 0 0 0 0 |
| − | 42 39 1 0 0 0 0 | + | 42 39 1 0 0 0 0 |
| − | 39 38 1 0 0 0 0 | + | 39 38 1 0 0 0 0 |
| − | 39 41 1 0 0 0 0 | + | 39 41 1 0 0 0 0 |
| − | 39 40 2 0 0 0 0 | + | 39 40 2 0 0 0 0 |
| − | S SKP 7 | + | S SKP 7 |
| − | ID BMCCPUAP0037 | + | ID BMCCPUAP0037 |
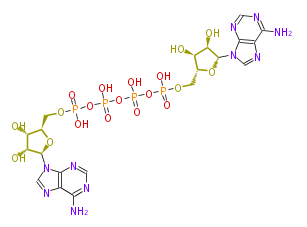
| − | NAME P1,P4-Bis(5'-adenosyl) tetraphosphate | + | NAME P1,P4-Bis(5'-adenosyl) tetraphosphate |
| − | FORMULA C20H28N10O19P4 | + | FORMULA C20H28N10O19P4 |
| − | EXACTMASS 836.0482 | + | EXACTMASS 836.0482 |
| − | AVERAGEMASS 836.3873 | + | AVERAGEMASS 836.3873 |
| − | SMILES OP(=O)(OP(OP(O)(=O)OP(O)(=O)OC[C@H]([C@H]6O)O[C@H]([C@@H]6O)n(c5)c(n4)c(n5)c(N)nc4)(O)=O)OC[C@H]([C@H]3O)O[C@H]([C@@H]3O)n(c2)c(n1)c(n2)c(N)nc1 | + | SMILES OP(=O)(OP(OP(O)(=O)OP(O)(=O)OC[C@H]([C@H]6O)O[C@H]([C@@H]6O)n(c5)c(n4)c(n5)c(N)nc4)(O)=O)OC[C@H]([C@H]3O)O[C@H]([C@@H]3O)n(c2)c(n1)c(n2)c(N)nc1 |
| − | KEGG http://www.genome.jp/dbget-bin/www_bget?compound+C01260 | + | KEGG http://www.genome.jp/dbget-bin/www_bget?compound+C01260 |
M END | M END | ||
| − | |||
Revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
53 58 0 0 1 0 0 0 0 0999 V2000
3.7321 2.1238 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
4.5981 1.6238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5981 0.6238 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
3.7321 0.1238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8660 0.6238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8660 1.6238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1229 -0.0454 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
2.5296 -0.9589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5241 -0.8544 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
2.0000 2.1238 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
4.1933 -1.5975 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.9854 -2.5757 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
4.8514 -3.0757 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
5.5945 -2.4065 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
6.5727 -2.6144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0718 -2.9824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9559 -4.0702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.1878 -1.4930 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
7.2418 -1.8713 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
19.9535 3.7612 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
19.0025 4.0702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
18.2593 3.4011 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
18.4672 2.4229 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
19.4183 2.1139 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
20.1614 2.7830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
19.4183 1.1139 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
18.4672 0.8049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
17.8795 1.6139 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
21.1614 2.7830 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
16.8795 1.6139 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
16.1363 2.2830 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
15.2703 1.7830 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
15.4782 0.8049 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
14.8091 0.0617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
16.2408 3.2775 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
14.3567 2.1898 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
16.4727 0.7003 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
13.8309 0.2696 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
13.1618 -0.4735 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0
13.9049 -1.1426 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
12.4186 0.1956 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
12.4927 -1.2167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
11.5145 -1.0087 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0
11.3066 -1.9869 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
11.7224 -0.0306 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
10.5364 -0.8008 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
9.8672 -1.5440 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0
10.6104 -2.2131 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
9.1241 -0.8748 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
9.1981 -2.2871 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
8.2200 -2.0792 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0
8.0120 -3.0574 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
8.4279 -1.1011 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4 3 1 0 0 0 0
3 2 2 0 0 0 0
2 1 1 0 0 0 0
1 6 2 0 0 0 0
6 5 1 0 0 0 0
4 5 2 0 0 0 0
5 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 4 1 0 0 0 0
6 10 1 0 0 0 0
11 12 1 0 0 0 0
12 13 1 0 0 0 0
13 14 1 0 0 0 0
14 18 1 6 0 0 0
11 18 1 6 0 0 0
12 16 1 1 0 0 0
13 17 1 1 0 0 0
14 15 1 0 0 0 0
15 19 1 0 0 0 0
11 9 1 0 0 0 0
23 22 1 0 0 0 0
22 21 2 0 0 0 0
21 20 1 0 0 0 0
20 25 2 0 0 0 0
25 24 1 0 0 0 0
23 24 2 0 0 0 0
24 26 1 0 0 0 0
26 27 2 0 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
25 29 1 0 0 0 0
30 31 1 0 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
33 37 1 6 0 0 0
30 37 1 6 0 0 0
31 35 1 1 0 0 0
32 36 1 1 0 0 0
33 34 1 0 0 0 0
34 38 1 0 0 0 0
30 28 1 0 0 0 0
19 51 1 0 0 0 0
50 51 1 0 0 0 0
51 52 1 0 0 0 0
51 53 2 0 0 0 0
50 47 1 0 0 0 0
47 46 1 0 0 0 0
47 49 1 0 0 0 0
47 48 2 0 0 0 0
46 43 1 0 0 0 0
43 42 1 0 0 0 0
43 45 1 0 0 0 0
43 44 2 0 0 0 0
42 39 1 0 0 0 0
39 38 1 0 0 0 0
39 41 1 0 0 0 0
39 40 2 0 0 0 0
S SKP 7
ID BMCCPUAP0037
NAME P1,P4-Bis(5'-adenosyl) tetraphosphate
FORMULA C20H28N10O19P4
EXACTMASS 836.0482
AVERAGEMASS 836.3873
SMILES OP(=O)(OP(OP(O)(=O)OP(O)(=O)OC[C@H]([C@H]6O)O[C@H]([C@@H]6O)n(c5)c(n4)c(n5)c(N)nc4)(O)=O)OC[C@H]([C@H]3O)O[C@H]([C@@H]3O)n(c2)c(n1)c(n2)c(N)nc1
KEGG http://www.genome.jp/dbget-bin/www_bget?compound+C01260
M END