Mol:BMCCPPPC0002
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 63 67 0 0 1 0 0 0 0 0999 V2000 | + | 63 67 0 0 1 0 0 0 0 0999 V2000 |
| − | 7.1714 2.6593 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.1714 2.6593 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.0913 3.2097 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 6.0913 3.2097 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 5.2340 2.3524 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 5.2340 2.3524 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 5.7844 1.2722 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.7844 1.2722 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.5893 0.2915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.5893 0.2915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.7844 -0.6893 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.7844 -0.6893 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2340 -1.7695 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 5.2340 -1.7695 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 6.0913 -2.6267 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 6.0913 -2.6267 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 7.1714 -2.0764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.1714 -2.0764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 8.1522 -2.2715 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 8.1522 -2.2715 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.1330 -2.0764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.1330 -2.0764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 10.2132 -2.6267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 10.2132 -2.6267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 11.0704 -1.7695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 11.0704 -1.7695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 10.5200 -0.6893 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 10.5200 -0.6893 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 10.7151 0.2915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 10.7151 0.2915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 10.5200 1.2722 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 10.5200 1.2722 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 11.0704 2.3524 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 11.0704 2.3524 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 10.2132 3.2097 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 10.2132 3.2097 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.1330 2.6593 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.1330 2.6593 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 8.1522 2.8544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 8.1522 2.8544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.3400 2.1037 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.3400 2.1037 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.3400 -1.5208 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.3400 -1.5208 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.9645 -1.5208 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.9645 -1.5208 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.9645 2.1037 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.9645 2.1037 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.5452 4.1007 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.5452 4.1007 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3841 3.9168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3841 3.9168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.6430 4.8827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.6430 4.8827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2463 2.5089 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2463 2.5089 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6170 1.7317 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6170 1.7317 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6293 1.8882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6293 1.8882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3430 -1.3155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3430 -1.3155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5269 -2.4766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5269 -2.4766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5610 -2.2178 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5610 -2.2178 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.9348 -3.6144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.9348 -3.6144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0012 -3.9728 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0012 -3.9728 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8448 -4.9605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8448 -4.9605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 10.3696 -3.6144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 10.3696 -3.6144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.5925 -4.2438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.5925 -4.2438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 12.0581 -1.9259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 12.0581 -1.9259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 12.4165 -2.8595 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 12.4165 -2.8595 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 13.4042 -3.0160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 13.4042 -3.0160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 13.2236 2.6935 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 13.2236 2.6935 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 13.5820 3.6270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 13.5820 3.6270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 14.5696 3.7835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 14.5696 3.7835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 10.3696 4.1974 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 10.3696 4.1974 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 11.3032 4.5557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 11.3032 4.5557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 8.1522 3.8544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 8.1522 3.8544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 14.0335 -2.2388 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 14.0335 -2.2388 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 13.7625 -3.9495 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 13.7625 -3.9495 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.6220 -5.5898 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.6220 -5.5898 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9112 -5.3189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9112 -5.3189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3022 -1.2519 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3022 -1.2519 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8539 -2.9249 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8539 -2.9249 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0000 1.1110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0000 1.1110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2710 2.8217 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2710 2.8217 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.6089 5.1415 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.6089 5.1415 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9359 5.5898 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9359 5.5898 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 8.6589 -3.8854 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 8.6589 -3.8854 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.7489 -5.2314 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.7489 -5.2314 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 15.1990 3.0063 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 15.1990 3.0063 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 14.9280 4.7171 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 14.9280 4.7171 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 12.0804 3.9264 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 12.0804 3.9264 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 11.4596 5.5434 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 11.4596 5.5434 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 21 4 1 0 0 0 0 | + | 21 4 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 3 2 1 0 0 0 0 | + | 3 2 1 0 0 0 0 |
| − | 1 21 1 0 0 0 0 | + | 1 21 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 1 20 2 0 0 0 0 | + | 1 20 2 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 7 32 1 6 0 0 0 | + | 7 32 1 6 0 0 0 |
| − | 2 26 1 6 0 0 0 | + | 2 26 1 6 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 8 34 1 1 0 0 0 | + | 8 34 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 41 48 2 0 0 0 0 | + | 41 48 2 0 0 0 0 |
| − | 14 23 1 0 0 0 0 | + | 14 23 1 0 0 0 0 |
| − | 41 49 1 0 0 0 0 | + | 41 49 1 0 0 0 0 |
| − | 17 42 1 0 0 0 0 | + | 17 42 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 36 50 1 0 0 0 0 | + | 36 50 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 36 51 2 0 0 0 0 | + | 36 51 2 0 0 0 0 |
| − | 23 11 1 0 0 0 0 | + | 23 11 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 18 19 2 0 0 0 0 | + | 18 19 2 0 0 0 0 |
| − | 33 52 1 0 0 0 0 | + | 33 52 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 33 53 2 0 0 0 0 | + | 33 53 2 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 24 16 1 0 0 0 0 | + | 24 16 1 0 0 0 0 |
| − | 30 54 1 0 0 0 0 | + | 30 54 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 30 55 2 0 0 0 0 | + | 30 55 2 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 19 24 1 0 0 0 0 | + | 19 24 1 0 0 0 0 |
| − | 27 56 1 0 0 0 0 | + | 27 56 1 0 0 0 0 |
| − | 16 15 1 0 0 0 0 | + | 16 15 1 0 0 0 0 |
| − | 27 57 2 0 0 0 0 | + | 27 57 2 0 0 0 0 |
| − | 15 14 1 0 0 0 0 | + | 15 14 1 0 0 0 0 |
| − | 12 37 1 0 0 0 0 | + | 12 37 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 11 10 1 0 0 0 0 | + | 11 10 1 0 0 0 0 |
| − | 38 58 1 0 0 0 0 | + | 38 58 1 0 0 0 0 |
| − | 10 9 2 0 0 0 0 | + | 10 9 2 0 0 0 0 |
| − | 38 59 2 0 0 0 0 | + | 38 59 2 0 0 0 0 |
| − | 6 22 2 0 0 0 0 | + | 6 22 2 0 0 0 0 |
| − | 7 31 1 1 0 0 0 | + | 7 31 1 1 0 0 0 |
| − | 20 19 1 0 0 0 0 | + | 20 19 1 0 0 0 0 |
| − | 3 28 1 1 0 0 0 | + | 3 28 1 1 0 0 0 |
| − | 44 60 1 0 0 0 0 | + | 44 60 1 0 0 0 0 |
| − | 9 8 1 0 0 0 0 | + | 9 8 1 0 0 0 0 |
| − | 44 61 2 0 0 0 0 | + | 44 61 2 0 0 0 0 |
| − | 8 7 1 0 0 0 0 | + | 8 7 1 0 0 0 0 |
| − | 18 45 1 0 0 0 0 | + | 18 45 1 0 0 0 0 |
| − | 7 6 1 0 0 0 0 | + | 7 6 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 22 9 1 0 0 0 0 | + | 22 9 1 0 0 0 0 |
| − | 46 62 1 0 0 0 0 | + | 46 62 1 0 0 0 0 |
| − | 2 1 1 0 0 0 0 | + | 2 1 1 0 0 0 0 |
| − | 46 63 2 0 0 0 0 | + | 46 63 2 0 0 0 0 |
| − | 13 39 1 0 0 0 0 | + | 13 39 1 0 0 0 0 |
| − | 2 25 1 1 0 0 0 | + | 2 25 1 1 0 0 0 |
| − | 20 47 1 0 0 0 0 | + | 20 47 1 0 0 0 0 |
| − | S SKP 7 | + | S SKP 7 |
| − | ID BMCCPPPC0002 | + | ID BMCCPPPC0002 |
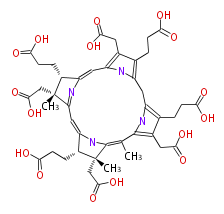
| − | NAME Precorrin 3A | + | NAME Precorrin 3A |
| − | FORMULA C43H50N4O16 | + | FORMULA C43H50N4O16 |
| − | EXACTMASS 878.3221 | + | EXACTMASS 878.3221 |
| − | AVERAGEMASS 878.8744 | + | AVERAGEMASS 878.8744 |
| − | SMILES C(O)(=O)CCc(c41)c(c(C(=C([C@@](C)5CC(O)=O)NC([C@H]5CCC(O)=O)=CC(=N2)[C@@]([C@H](CCC(O)=O)C2=Cc(n3)c(c(CCC(O)=O)c3C4)CC(O)=O)(CC(O)=O)C)C)n1)CC(O)=O | + | SMILES C(O)(=O)CCc(c41)c(c(C(=C([C@@](C)5CC(O)=O)NC([C@H]5CCC(O)=O)=CC(=N2)[C@@]([C@H](CCC(O)=O)C2=Cc(n3)c(c(CCC(O)=O)c3C4)CC(O)=O)(CC(O)=O)C)C)n1)CC(O)=O |
| − | KEGG http://www.genome.jp/dbget-bin/www_bget?compound+C05772 | + | KEGG http://www.genome.jp/dbget-bin/www_bget?compound+C05772 |
M END | M END | ||
| − | |||
Revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
63 67 0 0 1 0 0 0 0 0999 V2000
7.1714 2.6593 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.0913 3.2097 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
5.2340 2.3524 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
5.7844 1.2722 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.5893 0.2915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.7844 -0.6893 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.2340 -1.7695 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
6.0913 -2.6267 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
7.1714 -2.0764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
8.1522 -2.2715 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
9.1330 -2.0764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
10.2132 -2.6267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
11.0704 -1.7695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
10.5200 -0.6893 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
10.7151 0.2915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
10.5200 1.2722 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
11.0704 2.3524 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
10.2132 3.2097 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
9.1330 2.6593 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
8.1522 2.8544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.3400 2.1037 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
6.3400 -1.5208 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
9.9645 -1.5208 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
9.9645 2.1037 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
6.5452 4.1007 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.3841 3.9168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.6430 4.8827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2463 2.5089 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6170 1.7317 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6293 1.8882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3430 -1.3155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5269 -2.4766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5610 -2.2178 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.9348 -3.6144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.0012 -3.9728 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8448 -4.9605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
10.3696 -3.6144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
9.5925 -4.2438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
12.0581 -1.9259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
12.4165 -2.8595 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
13.4042 -3.0160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
13.2236 2.6935 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
13.5820 3.6270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
14.5696 3.7835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
10.3696 4.1974 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
11.3032 4.5557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
8.1522 3.8544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
14.0335 -2.2388 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
13.7625 -3.9495 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.6220 -5.5898 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9112 -5.3189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3022 -1.2519 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8539 -2.9249 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0000 1.1110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2710 2.8217 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.6089 5.1415 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9359 5.5898 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
8.6589 -3.8854 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
9.7489 -5.2314 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
15.1990 3.0063 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
14.9280 4.7171 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
12.0804 3.9264 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
11.4596 5.5434 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
21 4 1 0 0 0 0
4 3 1 0 0 0 0
3 2 1 0 0 0 0
1 21 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
1 20 2 0 0 0 0
39 40 1 0 0 0 0
7 32 1 6 0 0 0
2 26 1 6 0 0 0
40 41 1 0 0 0 0
8 34 1 1 0 0 0
34 35 1 0 0 0 0
41 48 2 0 0 0 0
14 23 1 0 0 0 0
41 49 1 0 0 0 0
17 42 1 0 0 0 0
11 12 2 0 0 0 0
35 36 1 0 0 0 0
12 13 1 0 0 0 0
36 50 1 0 0 0 0
13 14 2 0 0 0 0
36 51 2 0 0 0 0
23 11 1 0 0 0 0
32 33 1 0 0 0 0
18 19 2 0 0 0 0
33 52 1 0 0 0 0
28 29 1 0 0 0 0
33 53 2 0 0 0 0
42 43 1 0 0 0 0
29 30 1 0 0 0 0
24 16 1 0 0 0 0
30 54 1 0 0 0 0
16 17 2 0 0 0 0
30 55 2 0 0 0 0
17 18 1 0 0 0 0
26 27 1 0 0 0 0
19 24 1 0 0 0 0
27 56 1 0 0 0 0
16 15 1 0 0 0 0
27 57 2 0 0 0 0
15 14 1 0 0 0 0
12 37 1 0 0 0 0
43 44 1 0 0 0 0
37 38 1 0 0 0 0
11 10 1 0 0 0 0
38 58 1 0 0 0 0
10 9 2 0 0 0 0
38 59 2 0 0 0 0
6 22 2 0 0 0 0
7 31 1 1 0 0 0
20 19 1 0 0 0 0
3 28 1 1 0 0 0
44 60 1 0 0 0 0
9 8 1 0 0 0 0
44 61 2 0 0 0 0
8 7 1 0 0 0 0
18 45 1 0 0 0 0
7 6 1 0 0 0 0
45 46 1 0 0 0 0
22 9 1 0 0 0 0
46 62 1 0 0 0 0
2 1 1 0 0 0 0
46 63 2 0 0 0 0
13 39 1 0 0 0 0
2 25 1 1 0 0 0
20 47 1 0 0 0 0
S SKP 7
ID BMCCPPPC0002
NAME Precorrin 3A
FORMULA C43H50N4O16
EXACTMASS 878.3221
AVERAGEMASS 878.8744
SMILES C(O)(=O)CCc(c41)c(c(C(=C([C@@](C)5CC(O)=O)NC([C@H]5CCC(O)=O)=CC(=N2)[C@@]([C@H](CCC(O)=O)C2=Cc(n3)c(c(CCC(O)=O)c3C4)CC(O)=O)(CC(O)=O)C)C)n1)CC(O)=O
KEGG http://www.genome.jp/dbget-bin/www_bget?compound+C05772
M END