Mol:FLIAAAGS0011
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.5548 1.0095 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5548 1.0095 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0015 0.6884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0015 0.6884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5578 1.0095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5578 1.0095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1139 0.6885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1139 0.6885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1139 0.0225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1139 0.0225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6907 -0.3105 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6907 -0.3105 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2674 0.0225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2674 0.0225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2674 0.6885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2674 0.6885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6907 1.0215 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6907 1.0215 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0017 0.0464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0017 0.0464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5578 -0.2747 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5578 -0.2747 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6907 -0.9760 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6907 -0.9760 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8437 -0.3102 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8437 -0.3102 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8437 -0.9469 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8437 -0.9469 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3951 -1.2652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3951 -1.2652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9464 -0.9469 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9464 -0.9469 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9464 -0.3102 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9464 -0.3102 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3951 0.0081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3951 0.0081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4978 -1.2652 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4978 -1.2652 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6999 0.6408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6999 0.6408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3537 0.1837 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3537 0.1837 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8551 0.3776 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8551 0.3776 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3354 0.3719 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3354 0.3719 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7236 0.7325 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7236 0.7325 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2329 0.5496 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2329 0.5496 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5403 0.4156 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5403 0.4156 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8934 0.1575 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8934 0.1575 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5694 -0.1020 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5694 -0.1020 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5746 -0.6168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5746 -0.6168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1606 -1.0474 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1606 -1.0474 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1292 -0.8370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1292 -0.8370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5961 -0.4676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5961 -0.4676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1495 -0.6873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1495 -0.6873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5992 -0.4277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5992 -0.4277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0489 -0.6873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0489 -0.6873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0489 -1.2065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0489 -1.2065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5992 -1.4662 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5992 -1.4662 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1495 -1.2065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1495 -1.2065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4978 -1.4657 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4978 -1.4657 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5473 -0.9154 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5473 -0.9154 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4385 1.4662 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4385 1.4662 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7240 1.0537 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7240 1.0537 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 7 1 0 0 0 0 | + | 6 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 4 1 0 0 0 0 | + | 9 4 1 0 0 0 0 |
| − | 2 10 1 0 0 0 0 | + | 2 10 1 0 0 0 0 |
| − | 10 11 2 0 0 0 0 | + | 10 11 2 0 0 0 0 |
| − | 11 5 1 0 0 0 0 | + | 11 5 1 0 0 0 0 |
| − | 6 12 2 0 0 0 0 | + | 6 12 2 0 0 0 0 |
| − | 7 13 1 0 0 0 0 | + | 7 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 2 0 0 0 0 | + | 17 18 2 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 16 19 1 0 0 0 0 | + | 16 19 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 30 2 0 0 0 0 | + | 29 30 2 0 0 0 0 |
| − | 23 1 1 0 0 0 0 | + | 23 1 1 0 0 0 0 |
| − | 29 31 1 0 0 0 0 | + | 29 31 1 0 0 0 0 |
| − | 31 32 2 0 0 0 0 | + | 31 32 2 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 2 0 0 0 0 | + | 33 34 2 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 2 0 0 0 0 | + | 35 36 2 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 2 0 0 0 0 | + | 37 38 2 0 0 0 0 |
| − | 38 33 1 0 0 0 0 | + | 38 33 1 0 0 0 0 |
| − | 36 39 1 0 0 0 0 | + | 36 39 1 0 0 0 0 |
| − | 11 40 1 0 0 0 0 | + | 11 40 1 0 0 0 0 |
| − | 25 41 1 0 0 0 0 | + | 25 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 41 42 | + | M SAL 1 2 41 42 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 45 -6.4682 2.9603 | + | M SBV 1 45 -6.4682 2.9603 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FLIAAAGS0011 | + | ID FLIAAAGS0011 |
| − | KNApSAcK_ID C00010164 | + | KNApSAcK_ID C00010164 |
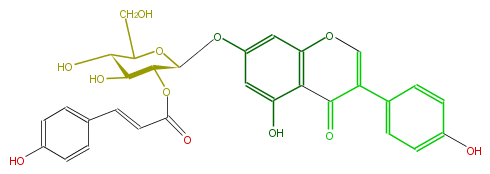
| − | NAME Genistein 7-O-(2''-p-coumaroylglucoside) | + | NAME Genistein 7-O-(2''-p-coumaroylglucoside) |
| − | CAS_RN 106915-84-8 | + | CAS_RN 106915-84-8 |
| − | FORMULA C30H26O12 | + | FORMULA C30H26O12 |
| − | EXACTMASS 578.1424262959999 | + | EXACTMASS 578.1424262959999 |
| − | AVERAGEMASS 578.5202400000001 | + | AVERAGEMASS 578.5202400000001 |
| − | SMILES C(O4)(C(OC(=O)C=Cc(c5)ccc(c5)O)C(C(C4CO)O)O)Oc(c1)cc(O3)c(C(C(=C3)c(c2)ccc(c2)O)=O)c1O | + | SMILES C(O4)(C(OC(=O)C=Cc(c5)ccc(c5)O)C(C(C4CO)O)O)Oc(c1)cc(O3)c(C(C(=C3)c(c2)ccc(c2)O)=O)c1O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
-0.5548 1.0095 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0015 0.6884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5578 1.0095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1139 0.6885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1139 0.0225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6907 -0.3105 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2674 0.0225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2674 0.6885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6907 1.0215 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0017 0.0464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5578 -0.2747 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6907 -0.9760 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8437 -0.3102 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8437 -0.9469 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3951 -1.2652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9464 -0.9469 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9464 -0.3102 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3951 0.0081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4978 -1.2652 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6999 0.6408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3537 0.1837 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8551 0.3776 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3354 0.3719 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7236 0.7325 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2329 0.5496 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5403 0.4156 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8934 0.1575 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5694 -0.1020 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5746 -0.6168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1606 -1.0474 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1292 -0.8370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5961 -0.4676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1495 -0.6873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5992 -0.4277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0489 -0.6873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0489 -1.2065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5992 -1.4662 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1495 -1.2065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4978 -1.4657 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5473 -0.9154 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4385 1.4662 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7240 1.0537 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 4 1 0 0 0 0
2 10 1 0 0 0 0
10 11 2 0 0 0 0
11 5 1 0 0 0 0
6 12 2 0 0 0 0
7 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 17 1 0 0 0 0
17 18 2 0 0 0 0
18 13 1 0 0 0 0
16 19 1 0 0 0 0
20 21 1 1 0 0 0
21 22 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 20 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
28 29 1 0 0 0 0
29 30 2 0 0 0 0
23 1 1 0 0 0 0
29 31 1 0 0 0 0
31 32 2 0 0 0 0
32 33 1 0 0 0 0
33 34 2 0 0 0 0
34 35 1 0 0 0 0
35 36 2 0 0 0 0
36 37 1 0 0 0 0
37 38 2 0 0 0 0
38 33 1 0 0 0 0
36 39 1 0 0 0 0
11 40 1 0 0 0 0
25 41 1 0 0 0 0
41 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 41 42
M SBL 1 1 45
M SMT 1 ^CH2OH
M SBV 1 45 -6.4682 2.9603
S SKP 8
ID FLIAAAGS0011
KNApSAcK_ID C00010164
NAME Genistein 7-O-(2''-p-coumaroylglucoside)
CAS_RN 106915-84-8
FORMULA C30H26O12
EXACTMASS 578.1424262959999
AVERAGEMASS 578.5202400000001
SMILES C(O4)(C(OC(=O)C=Cc(c5)ccc(c5)O)C(C(C4CO)O)O)Oc(c1)cc(O3)c(C(C(=C3)c(c2)ccc(c2)O)=O)c1O
M END