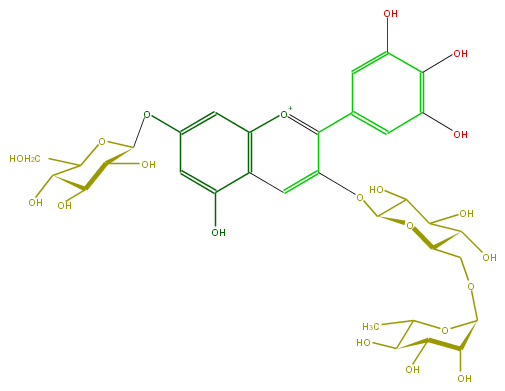
Mol:FL7AAGGL0013
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 54 59 0 0 0 0 0 0 0 0999 V2000 | + | 54 59 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.2837 1.4401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2837 1.4401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2836 0.5159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2836 0.5159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4832 0.0538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4832 0.0538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3171 0.5159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3171 0.5159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3170 1.4401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3170 1.4401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4832 1.9023 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4832 1.9023 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1176 0.0538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1176 0.0538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9180 0.5159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9180 0.5159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9180 1.4401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9180 1.4401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1175 1.9023 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 | + | 1.1175 1.9023 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7181 1.9021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7181 1.9021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5339 1.4311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5339 1.4311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3495 1.9020 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3495 1.9020 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3495 2.8440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3495 2.8440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5339 3.3151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5339 3.3151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7179 2.8440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7179 2.8440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0838 1.9021 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0838 1.9021 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1650 3.3149 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1650 3.3149 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4832 -0.8700 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4832 -0.8700 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8991 -0.0511 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8991 -0.0511 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5339 4.2568 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5339 4.2568 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1399 -2.9371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1399 -2.9371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7541 -3.6056 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7541 -3.6056 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4961 -3.3933 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4961 -3.3933 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2427 -3.6056 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2427 -3.6056 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.6288 -2.9371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.6288 -2.9371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8865 -3.1491 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8865 -3.1491 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2795 -3.0552 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2795 -3.0552 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9732 -0.1195 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9732 -0.1195 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3049 -0.5055 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3049 -0.5055 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0536 -0.6930 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0536 -0.6930 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5941 -1.2498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5941 -1.2498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2627 -0.8639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2627 -0.8639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5137 -0.6765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5137 -0.6765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3514 0.1385 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3514 0.1385 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2980 -0.4243 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2980 -0.4243 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.8331 -1.4416 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.8331 -1.4416 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2288 -1.4609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2288 -1.4609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4783 -2.1282 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4783 -2.1282 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0604 -3.4204 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0604 -3.4204 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0447 -4.0598 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0447 -4.0598 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2427 -4.2568 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2427 -4.2568 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1650 1.4312 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1650 1.4312 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2752 0.4442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2752 0.4442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5150 0.1238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5150 0.1238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0333 0.7240 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0333 0.7240 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3815 1.0908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3815 1.0908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1246 1.2608 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1246 1.2608 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6278 0.6664 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6278 0.6664 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7538 -0.1585 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7538 -0.1585 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0847 -0.2372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0847 -0.2372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1272 0.7268 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1272 0.7268 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4841 0.8692 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4841 0.8692 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.8331 0.3691 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.8331 0.3691 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 1 17 1 0 0 0 0 | + | 1 17 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 20 8 1 0 0 0 0 | + | 20 8 1 0 0 0 0 |
| − | 15 21 1 0 0 0 0 | + | 15 21 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 29 1 0 0 0 0 | + | 34 29 1 0 0 0 0 |
| − | 29 35 1 0 0 0 0 | + | 29 35 1 0 0 0 0 |
| − | 34 36 1 0 0 0 0 | + | 34 36 1 0 0 0 0 |
| − | 33 37 1 0 0 0 0 | + | 33 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 26 39 1 0 0 0 0 | + | 26 39 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 23 40 1 0 0 0 0 | + | 23 40 1 0 0 0 0 |
| − | 24 41 1 0 0 0 0 | + | 24 41 1 0 0 0 0 |
| − | 25 42 1 0 0 0 0 | + | 25 42 1 0 0 0 0 |
| − | 30 20 1 0 0 0 0 | + | 30 20 1 0 0 0 0 |
| − | 13 43 1 0 0 0 0 | + | 13 43 1 0 0 0 0 |
| − | 44 45 1 1 0 0 0 | + | 44 45 1 1 0 0 0 |
| − | 45 46 1 1 0 0 0 | + | 45 46 1 1 0 0 0 |
| − | 47 46 1 1 0 0 0 | + | 47 46 1 1 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 49 44 1 0 0 0 0 | + | 49 44 1 0 0 0 0 |
| − | 44 50 1 0 0 0 0 | + | 44 50 1 0 0 0 0 |
| − | 45 51 1 0 0 0 0 | + | 45 51 1 0 0 0 0 |
| − | 46 52 1 0 0 0 0 | + | 46 52 1 0 0 0 0 |
| − | 47 17 1 0 0 0 0 | + | 47 17 1 0 0 0 0 |
| − | 53 54 1 0 0 0 0 | + | 53 54 1 0 0 0 0 |
| − | 49 53 1 0 0 0 0 | + | 49 53 1 0 0 0 0 |
| − | M CHG 1 10 1 | + | M CHG 1 10 1 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 53 54 | + | M SAL 1 2 53 54 |
| − | M SBL 1 1 59 | + | M SBL 1 1 59 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 59 0.8562 -0.2027 | + | M SBV 1 59 0.8562 -0.2027 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL7AAGGL0013 | + | ID FL7AAGGL0013 |
| − | FORMULA C33H41O21 | + | FORMULA C33H41O21 |
| − | EXACTMASS 773.214033374 | + | EXACTMASS 773.214033374 |
| − | AVERAGEMASS 773.66604 | + | AVERAGEMASS 773.66604 |
| − | SMILES C(O1)(CO)C(C(C(C1Oc(c2)cc(O)c(c4)c2[o+1]c(c4OC(O5)C(C(O)C(C5COC(C6O)OC(C(O)C(O)6)C)O)O)c(c3)cc(O)c(O)c(O)3)O)O)O | + | SMILES C(O1)(CO)C(C(C(C1Oc(c2)cc(O)c(c4)c2[o+1]c(c4OC(O5)C(C(O)C(C5COC(C6O)OC(C(O)C(O)6)C)O)O)c(c3)cc(O)c(O)c(O)3)O)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
54 59 0 0 0 0 0 0 0 0999 V2000
-1.2837 1.4401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2836 0.5159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4832 0.0538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3171 0.5159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3170 1.4401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4832 1.9023 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1176 0.0538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9180 0.5159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9180 1.4401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1175 1.9023 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
2.7181 1.9021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5339 1.4311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3495 1.9020 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3495 2.8440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5339 3.3151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7179 2.8440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0838 1.9021 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.1650 3.3149 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4832 -0.8700 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8991 -0.0511 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5339 4.2568 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1399 -2.9371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7541 -3.6056 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4961 -3.3933 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.2427 -3.6056 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.6288 -2.9371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8865 -3.1491 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2795 -3.0552 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9732 -0.1195 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3049 -0.5055 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0536 -0.6930 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5941 -1.2498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.2627 -0.8639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5137 -0.6765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3514 0.1385 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.2980 -0.4243 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.8331 -1.4416 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.2288 -1.4609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.4783 -2.1282 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0604 -3.4204 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0447 -4.0598 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.2427 -4.2568 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.1650 1.4312 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2752 0.4442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5150 0.1238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0333 0.7240 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3815 1.0908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1246 1.2608 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6278 0.6664 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7538 -0.1585 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0847 -0.2372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1272 0.7268 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.4841 0.8692 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.8331 0.3691 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
1 17 1 0 0 0 0
14 18 1 0 0 0 0
3 19 1 0 0 0 0
20 8 1 0 0 0 0
15 21 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
29 30 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 29 1 0 0 0 0
29 35 1 0 0 0 0
34 36 1 0 0 0 0
33 37 1 0 0 0 0
32 38 1 0 0 0 0
26 39 1 0 0 0 0
38 39 1 0 0 0 0
23 40 1 0 0 0 0
24 41 1 0 0 0 0
25 42 1 0 0 0 0
30 20 1 0 0 0 0
13 43 1 0 0 0 0
44 45 1 1 0 0 0
45 46 1 1 0 0 0
47 46 1 1 0 0 0
47 48 1 0 0 0 0
48 49 1 0 0 0 0
49 44 1 0 0 0 0
44 50 1 0 0 0 0
45 51 1 0 0 0 0
46 52 1 0 0 0 0
47 17 1 0 0 0 0
53 54 1 0 0 0 0
49 53 1 0 0 0 0
M CHG 1 10 1
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 53 54
M SBL 1 1 59
M SMT 1 ^ CH2OH
M SBV 1 59 0.8562 -0.2027
S SKP 5
ID FL7AAGGL0013
FORMULA C33H41O21
EXACTMASS 773.214033374
AVERAGEMASS 773.66604
SMILES C(O1)(CO)C(C(C(C1Oc(c2)cc(O)c(c4)c2[o+1]c(c4OC(O5)C(C(O)C(C5COC(C6O)OC(C(O)C(O)6)C)O)O)c(c3)cc(O)c(O)c(O)3)O)O)O
M END