Mol:FL7AACGL0065
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 60 65 0 0 0 0 0 0 0 0999 V2000 | + | 60 65 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.5510 1.3250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5510 1.3250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5510 0.6827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5510 0.6827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9947 0.3615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9947 0.3615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4384 0.6827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4384 0.6827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4384 1.3250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4384 1.3250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9947 1.6462 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9947 1.6462 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8821 0.3615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8821 0.3615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3258 0.6827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3258 0.6827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3258 1.3250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3258 1.3250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8821 1.6462 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 | + | -0.8821 1.6462 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2303 1.6461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2303 1.6461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7973 1.3188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7973 1.3188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3643 1.6461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3643 1.6461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3643 2.3008 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3643 2.3008 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7973 2.6281 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7973 2.6281 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2303 2.3008 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2303 2.3008 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1071 1.6461 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1071 1.6461 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9311 2.6280 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9311 2.6280 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9947 -0.2806 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9947 -0.2806 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0903 -0.0380 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0903 -0.0380 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1894 0.2586 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 1.1894 0.2586 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 1.5911 -0.2717 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.5911 -0.2717 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.1696 -0.0467 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.1696 -0.0467 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.7277 -0.0407 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.7277 -0.0407 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.3221 0.3650 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.3221 0.3650 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.8161 0.0979 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8161 0.0979 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9649 -0.3021 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9649 -0.3021 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5010 -0.6031 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5010 -0.6031 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7189 -0.1728 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7189 -0.1728 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7973 3.2826 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7973 3.2826 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1535 -0.5548 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -4.1535 -0.5548 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -3.7823 -1.0448 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.7823 -1.0448 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -3.2478 -0.8369 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.2478 -0.8369 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.7320 -0.8313 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.7320 -0.8313 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -3.1068 -0.4565 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1068 -0.4565 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5744 -0.7033 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -3.5744 -0.7033 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -4.6672 -0.2582 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6672 -0.2582 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1327 -1.5060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1327 -1.5060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9415 -1.3510 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9415 -1.3510 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9450 -0.1281 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9450 -0.1281 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6063 0.2756 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6063 0.2756 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3221 0.3650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3221 0.3650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1388 1.0070 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1388 1.0070 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6745 1.2838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6745 1.2838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2749 0.9371 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2749 0.9371 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6745 1.9235 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6745 1.9235 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1063 2.1728 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1063 2.1728 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1063 2.8526 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1063 2.8526 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2959 -2.4876 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 0.2959 -2.4876 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 0.8780 -2.3374 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.8780 -2.3374 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.8503 -1.7427 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.8503 -1.7427 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.1409 -1.2295 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.1409 -1.2295 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.5024 -1.4548 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5024 -1.4548 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5588 -2.0745 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 0.5588 -2.0745 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.2959 -2.9604 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2959 -2.9604 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8343 -1.8268 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8343 -1.8268 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6673 1.8490 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6673 1.8490 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0757 -2.1167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0757 -2.1167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3656 -1.7465 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3656 -1.7465 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6247 -3.2826 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6247 -3.2826 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 1 17 1 0 0 0 0 | + | 1 17 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 20 8 1 0 0 0 0 | + | 20 8 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 22 27 1 0 0 0 0 | + | 22 27 1 0 0 0 0 |
| − | 23 28 1 0 0 0 0 | + | 23 28 1 0 0 0 0 |
| − | 24 29 1 0 0 0 0 | + | 24 29 1 0 0 0 0 |
| − | 15 30 1 0 0 0 0 | + | 15 30 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 34 19 1 0 0 0 0 | + | 34 19 1 0 0 0 0 |
| − | 25 42 1 0 0 0 0 | + | 25 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 44 45 2 0 0 0 0 | + | 44 45 2 0 0 0 0 |
| − | 44 46 1 0 0 0 0 | + | 44 46 1 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 2 0 0 0 0 | + | 47 48 2 0 0 0 0 |
| − | 44 43 1 0 0 0 0 | + | 44 43 1 0 0 0 0 |
| − | 49 50 1 1 0 0 0 | + | 49 50 1 1 0 0 0 |
| − | 50 51 1 1 0 0 0 | + | 50 51 1 1 0 0 0 |
| − | 52 51 1 1 0 0 0 | + | 52 51 1 1 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 53 54 1 0 0 0 0 | + | 53 54 1 0 0 0 0 |
| − | 54 49 1 0 0 0 0 | + | 54 49 1 0 0 0 0 |
| − | 49 55 1 0 0 0 0 | + | 49 55 1 0 0 0 0 |
| − | 20 21 1 0 0 0 0 | + | 20 21 1 0 0 0 0 |
| − | 27 52 1 0 0 0 0 | + | 27 52 1 0 0 0 0 |
| − | 56 51 1 0 0 0 0 | + | 56 51 1 0 0 0 0 |
| − | 47 57 1 0 0 0 0 | + | 47 57 1 0 0 0 0 |
| − | 54 58 1 0 0 0 0 | + | 54 58 1 0 0 0 0 |
| − | 58 59 1 0 0 0 0 | + | 58 59 1 0 0 0 0 |
| − | 50 60 1 0 0 0 0 | + | 50 60 1 0 0 0 0 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 58 59 | + | M SAL 3 2 58 59 |
| − | M SBL 3 1 63 | + | M SBL 3 1 63 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SVB 3 63 0.0903 -2.2 | + | M SVB 3 63 0.0903 -2.2 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 40 41 | + | M SAL 2 2 40 41 |
| − | M SBL 2 1 42 | + | M SBL 2 1 42 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 42 -4.0393 -0.2012 | + | M SVB 2 42 -4.0393 -0.2012 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 3 47 48 57 | + | M SAL 1 3 47 48 57 |
| − | M SBL 1 1 49 | + | M SBL 1 1 49 |
| − | M SMT 1 COOH | + | M SMT 1 COOH |
| − | M SVB 1 49 4.1063 2.1728 | + | M SVB 1 49 4.1063 2.1728 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL7AACGL0065 | + | ID FL7AACGL0065 |
| − | KNApSAcK_ID C00006841 | + | KNApSAcK_ID C00006841 |
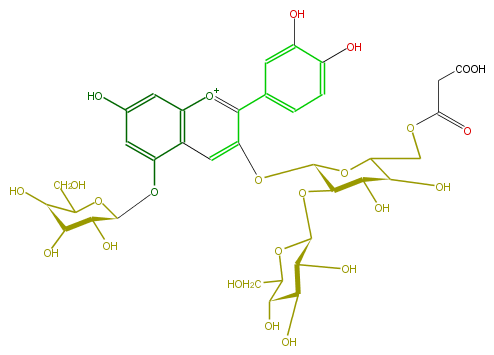
| − | NAME Cyanidin 3-(6''-malonylsophoroside)-5-glucoside | + | NAME Cyanidin 3-(6''-malonylsophoroside)-5-glucoside |
| − | CAS_RN 63003-09-8,117829-27-3 | + | CAS_RN 63003-09-8,117829-27-3 |
| − | FORMULA C36H43O24 | + | FORMULA C36H43O24 |
| − | EXACTMASS 859.2144273040001 | + | EXACTMASS 859.2144273040001 |
| − | AVERAGEMASS 859.71222 | + | AVERAGEMASS 859.71222 |
| − | SMILES [C@H]([C@H](Oc(c24)cc(O)cc2[o+1]c(c(O[C@H]([C@H](O[C@@H]([C@@H](O)6)OC(CO)[C@H](O)[C@H](O)6)5)OC(COC(=O)CC(O)=O)[C@@H](O)[C@H]5O)c4)c(c3)ccc(O)c3O)1)([C@@H](O)[C@H](C(CO)O1)O)O | + | SMILES [C@H]([C@H](Oc(c24)cc(O)cc2[o+1]c(c(O[C@H]([C@H](O[C@@H]([C@@H](O)6)OC(CO)[C@H](O)[C@H](O)6)5)OC(COC(=O)CC(O)=O)[C@@H](O)[C@H]5O)c4)c(c3)ccc(O)c3O)1)([C@@H](O)[C@H](C(CO)O1)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
60 65 0 0 0 0 0 0 0 0999 V2000
-2.5510 1.3250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5510 0.6827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9947 0.3615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4384 0.6827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4384 1.3250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9947 1.6462 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8821 0.3615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3258 0.6827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3258 1.3250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8821 1.6462 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
0.2303 1.6461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7973 1.3188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3643 1.6461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3643 2.3008 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7973 2.6281 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2303 2.3008 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1071 1.6461 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9311 2.6280 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9947 -0.2806 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0903 -0.0380 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1894 0.2586 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
1.5911 -0.2717 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.1696 -0.0467 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.7277 -0.0407 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.3221 0.3650 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.8161 0.0979 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9649 -0.3021 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5010 -0.6031 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7189 -0.1728 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7973 3.2826 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1535 -0.5548 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-3.7823 -1.0448 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-3.2478 -0.8369 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.7320 -0.8313 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-3.1068 -0.4565 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5744 -0.7033 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-4.6672 -0.2582 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1327 -1.5060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9415 -1.3510 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9450 -0.1281 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6063 0.2756 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3221 0.3650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1388 1.0070 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6745 1.2838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2749 0.9371 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6745 1.9235 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1063 2.1728 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1063 2.8526 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2959 -2.4876 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
0.8780 -2.3374 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.8503 -1.7427 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.1409 -1.2295 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.5024 -1.4548 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5588 -2.0745 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.2959 -2.9604 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8343 -1.8268 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6673 1.8490 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0757 -2.1167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3656 -1.7465 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6247 -3.2826 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
1 17 1 0 0 0 0
14 18 1 0 0 0 0
3 19 1 0 0 0 0
20 8 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
22 27 1 0 0 0 0
23 28 1 0 0 0 0
24 29 1 0 0 0 0
15 30 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
36 40 1 0 0 0 0
40 41 1 0 0 0 0
34 19 1 0 0 0 0
25 42 1 0 0 0 0
42 43 1 0 0 0 0
44 45 2 0 0 0 0
44 46 1 0 0 0 0
46 47 1 0 0 0 0
47 48 2 0 0 0 0
44 43 1 0 0 0 0
49 50 1 1 0 0 0
50 51 1 1 0 0 0
52 51 1 1 0 0 0
52 53 1 0 0 0 0
53 54 1 0 0 0 0
54 49 1 0 0 0 0
49 55 1 0 0 0 0
20 21 1 0 0 0 0
27 52 1 0 0 0 0
56 51 1 0 0 0 0
47 57 1 0 0 0 0
54 58 1 0 0 0 0
58 59 1 0 0 0 0
50 60 1 0 0 0 0
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 58 59
M SBL 3 1 63
M SMT 3 CH2OH
M SVB 3 63 0.0903 -2.2
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 40 41
M SBL 2 1 42
M SMT 2 CH2OH
M SVB 2 42 -4.0393 -0.2012
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 3 47 48 57
M SBL 1 1 49
M SMT 1 COOH
M SVB 1 49 4.1063 2.1728
S SKP 8
ID FL7AACGL0065
KNApSAcK_ID C00006841
NAME Cyanidin 3-(6''-malonylsophoroside)-5-glucoside
CAS_RN 63003-09-8,117829-27-3
FORMULA C36H43O24
EXACTMASS 859.2144273040001
AVERAGEMASS 859.71222
SMILES [C@H]([C@H](Oc(c24)cc(O)cc2[o+1]c(c(O[C@H]([C@H](O[C@@H]([C@@H](O)6)OC(CO)[C@H](O)[C@H](O)6)5)OC(COC(=O)CC(O)=O)[C@@H](O)[C@H]5O)c4)c(c3)ccc(O)c3O)1)([C@@H](O)[C@H](C(CO)O1)O)O
M END