Mol:FL5FGLNS0012
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 31 33 0 0 0 0 0 0 0 0999 V2000 | + | 31 33 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.0211 -1.8725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0211 -1.8725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2408 -1.2688 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2408 -1.2688 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8734 -1.1573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8734 -1.1573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2863 -1.6494 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2863 -1.6494 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0666 -2.2530 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0666 -2.2530 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4340 -2.3646 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4340 -2.3646 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9189 -1.5378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9189 -1.5378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3318 -2.0299 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3318 -2.0299 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1121 -2.6336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1121 -2.6336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4795 -2.7451 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4795 -2.7451 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0902 -1.0672 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0902 -1.0672 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5249 -3.1254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5249 -3.1254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1695 -3.0117 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1695 -3.0117 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.5904 -3.5132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.5904 -3.5132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3665 -4.1284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3665 -4.1284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7217 -4.2422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7217 -4.2422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3009 -3.7406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3009 -3.7406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3887 -1.9840 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3887 -1.9840 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0930 -0.5539 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0930 -0.5539 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4992 -2.9981 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4992 -2.9981 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6017 -3.9929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6017 -3.9929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7536 -4.1999 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7536 -4.1999 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9875 -4.8427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9875 -4.8427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3166 -1.8562 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3166 -1.8562 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.3014 -1.6826 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.3014 -1.6826 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.7871 -4.6299 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.7871 -4.6299 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.5996 -4.4866 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.5996 -4.4866 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.5752 -3.3395 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.5752 -3.3395 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -7.5600 -3.1659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -7.5600 -3.1659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5213 -1.2349 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5213 -1.2349 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2966 -0.9528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2966 -0.9528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 6 20 1 0 0 0 0 | + | 6 20 1 0 0 0 0 |
| − | 20 21 1 0 0 0 0 | + | 20 21 1 0 0 0 0 |
| − | 17 22 1 0 0 0 0 | + | 17 22 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 8 24 1 0 0 0 0 | + | 8 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 15 26 1 0 0 0 0 | + | 15 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 14 28 1 0 0 0 0 | + | 14 28 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 2 30 1 0 0 0 0 | + | 2 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | M STY 1 6 SUP | + | M STY 1 6 SUP |
| − | M SLB 1 6 6 | + | M SLB 1 6 6 |
| − | M SAL 6 2 30 31 | + | M SAL 6 2 30 31 |
| − | M SBL 6 1 32 | + | M SBL 6 1 32 |
| − | M SMT 6 OCH3 | + | M SMT 6 OCH3 |
| − | M SVB 6 32 -2.9555 -0.5453 | + | M SVB 6 32 -2.9555 -0.5453 |
| − | M STY 1 5 SUP | + | M STY 1 5 SUP |
| − | M SLB 1 5 5 | + | M SLB 1 5 5 |
| − | M SAL 5 2 28 29 | + | M SAL 5 2 28 29 |
| − | M SBL 5 1 30 | + | M SBL 5 1 30 |
| − | M SMT 5 OCH3 | + | M SMT 5 OCH3 |
| − | M SVB 5 30 1.8838 0.1204 | + | M SVB 5 30 1.8838 0.1204 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 26 27 | + | M SAL 4 2 26 27 |
| − | M SBL 4 1 28 | + | M SBL 4 1 28 |
| − | M SMT 4 OCH3 | + | M SMT 4 OCH3 |
| − | M SVB 4 28 2.241 1.3086 | + | M SVB 4 28 2.241 1.3086 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 24 25 | + | M SAL 3 2 24 25 |
| − | M SBL 3 1 26 | + | M SBL 3 1 26 |
| − | M SMT 3 OCH3 | + | M SMT 3 OCH3 |
| − | M SVB 3 26 0.183 -0.843 | + | M SVB 3 26 0.183 -0.843 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 22 23 | + | M SAL 2 2 22 23 |
| − | M SBL 2 1 24 | + | M SBL 2 1 24 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 24 0.183 1.6001 | + | M SVB 2 24 0.183 1.6001 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 20 21 | + | M SAL 1 2 20 21 |
| − | M SBL 1 1 22 | + | M SBL 1 1 22 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 22 -1.4067 0.8998 | + | M SVB 1 22 -1.4067 0.8998 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FGLNS0012 | + | ID FL5FGLNS0012 |
| − | KNApSAcK_ID C00004856 | + | KNApSAcK_ID C00004856 |
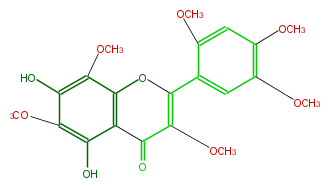
| − | NAME 5,7-Dihydroxy-3,6,8,2',4',5'-hexamethoxyflavone | + | NAME 5,7-Dihydroxy-3,6,8,2',4',5'-hexamethoxyflavone |
| − | CAS_RN 113744-04-0 | + | CAS_RN 113744-04-0 |
| − | FORMULA C21H22O10 | + | FORMULA C21H22O10 |
| − | EXACTMASS 434.121296924 | + | EXACTMASS 434.121296924 |
| − | AVERAGEMASS 434.39338 | + | AVERAGEMASS 434.39338 |
| − | SMILES COc(c1)c(C(O3)=C(C(c(c32)c(c(c(O)c2OC)OC)O)=O)OC)cc(OC)c1OC | + | SMILES COc(c1)c(C(O3)=C(C(c(c32)c(c(c(O)c2OC)OC)O)=O)OC)cc(OC)c1OC |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/ 31 33 0 0 0 0 0 0 0 0999 V2000 -2.0211 -1.8725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -2.2408 -1.2688 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -2.8734 -1.1573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -3.2863 -1.6494 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -3.0666 -2.2530 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -2.4340 -2.3646 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -3.9189 -1.5378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -4.3318 -2.0299 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -4.1121 -2.6336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -3.4795 -2.7451 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -4.0902 -1.0672 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -4.5249 -3.1254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -5.1695 -3.0117 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -5.5904 -3.5132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -5.3665 -4.1284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -4.7217 -4.2422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -4.3009 -3.7406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -1.3887 -1.9840 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -3.0930 -0.5539 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -2.4992 -2.9981 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -2.6017 -3.9929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -3.7536 -4.1999 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -2.9875 -4.8427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -5.3166 -1.8562 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -6.3014 -1.6826 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -5.7871 -4.6299 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -6.5996 -4.4866 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -6.5752 -3.3395 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -7.5600 -3.1659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -1.5213 -1.2349 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -2.2966 -0.9528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 1 2 1 0 0 0 0 2 3 2 0 0 0 0 3 4 1 0 0 0 0 4 5 2 0 0 0 0 5 6 1 0 0 0 0 6 1 2 0 0 0 0 4 7 1 0 0 0 0 7 8 1 0 0 0 0 8 9 2 0 0 0 0 9 10 1 0 0 0 0 10 5 1 0 0 0 0 7 11 2 0 0 0 0 9 12 1 0 0 0 0 12 13 2 0 0 0 0 13 14 1 0 0 0 0 14 15 2 0 0 0 0 15 16 1 0 0 0 0 16 17 2 0 0 0 0 17 12 1 0 0 0 0 1 18 1 0 0 0 0 3 19 1 0 0 0 0 6 20 1 0 0 0 0 20 21 1 0 0 0 0 17 22 1 0 0 0 0 22 23 1 0 0 0 0 8 24 1 0 0 0 0 24 25 1 0 0 0 0 15 26 1 0 0 0 0 26 27 1 0 0 0 0 14 28 1 0 0 0 0 28 29 1 0 0 0 0 2 30 1 0 0 0 0 30 31 1 0 0 0 0 M STY 1 6 SUP M SLB 1 6 6 M SAL 6 2 30 31 M SBL 6 1 32 M SMT 6 OCH3 M SVB 6 32 -2.9555 -0.5453 M STY 1 5 SUP M SLB 1 5 5 M SAL 5 2 28 29 M SBL 5 1 30 M SMT 5 OCH3 M SVB 5 30 1.8838 0.1204 M STY 1 4 SUP M SLB 1 4 4 M SAL 4 2 26 27 M SBL 4 1 28 M SMT 4 OCH3 M SVB 4 28 2.241 1.3086 M STY 1 3 SUP M SLB 1 3 3 M SAL 3 2 24 25 M SBL 3 1 26 M SMT 3 OCH3 M SVB 3 26 0.183 -0.843 M STY 1 2 SUP M SLB 1 2 2 M SAL 2 2 22 23 M SBL 2 1 24 M SMT 2 OCH3 M SVB 2 24 0.183 1.6001 M STY 1 1 SUP M SLB 1 1 1 M SAL 1 2 20 21 M SBL 1 1 22 M SMT 1 OCH3 M SVB 1 22 -1.4067 0.8998 S SKP 8 ID FL5FGLNS0012 KNApSAcK_ID C00004856 NAME 5,7-Dihydroxy-3,6,8,2',4',5'-hexamethoxyflavone CAS_RN 113744-04-0 FORMULA C21H22O10 EXACTMASS 434.121296924 AVERAGEMASS 434.39338 SMILES COc(c1)c(C(O3)=C(C(c(c32)c(c(c(O)c2OC)OC)O)=O)OC)cc(OC)c1OC M END