Mol:FL5FFAGS0012
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.6310 -0.6427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6310 -0.6427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6310 -1.4676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6310 -1.4676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9166 -1.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9166 -1.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2021 -1.4676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2021 -1.4676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2021 -0.6427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2021 -0.6427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9166 -0.2302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9166 -0.2302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5123 -1.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5123 -1.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2268 -1.4676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2268 -1.4676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2268 -0.6427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2268 -0.6427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5123 -0.2302 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5123 -0.2302 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5123 -2.5235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5123 -2.5235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1263 -0.0715 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1263 -0.0715 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8545 -0.4918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8545 -0.4918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5827 -0.0715 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5827 -0.0715 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5827 0.7694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5827 0.7694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8545 1.1899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8545 1.1899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1263 0.7694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1263 0.7694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9166 -2.5344 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9166 -2.5344 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3620 1.2194 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3620 1.2194 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9502 -1.9937 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9502 -1.9937 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4845 -0.1499 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4845 -0.1499 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9166 0.5944 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9166 0.5944 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7701 2.4415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7701 2.4415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9025 1.6204 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9025 1.6204 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2139 1.3290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2139 1.3290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7105 0.7197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7105 0.7197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5517 1.5409 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5517 1.5409 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3463 1.6998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3463 1.6998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3994 2.5672 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3994 2.5672 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2588 2.0109 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2588 2.0109 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0787 0.4355 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0787 0.4355 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5883 -2.1406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5883 -2.1406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8731 -1.5579 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8731 -1.5579 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3193 -0.8822 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3193 -0.8822 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3233 0.0050 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3233 0.0050 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7971 -0.6844 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7971 -0.6844 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3764 -1.4255 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3764 -1.4255 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2621 -2.7532 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2621 -2.7532 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9693 -2.3913 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9693 -2.3913 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0240 -1.9801 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0240 -1.9801 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3620 -0.9422 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3620 -0.9422 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3782 2.7532 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3782 2.7532 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4607 2.2234 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4607 2.2234 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 20 8 1 0 0 0 0 | + | 20 8 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 1 21 1 0 0 0 0 | + | 1 21 1 0 0 0 0 |
| − | 6 22 1 0 0 0 0 | + | 6 22 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 25 31 1 0 0 0 0 | + | 25 31 1 0 0 0 0 |
| − | 22 26 1 0 0 0 0 | + | 22 26 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 37 40 1 0 0 0 0 | + | 37 40 1 0 0 0 0 |
| − | 35 21 1 0 0 0 0 | + | 35 21 1 0 0 0 0 |
| − | 36 41 1 0 0 0 0 | + | 36 41 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 28 42 1 0 0 0 0 | + | 28 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 42 43 | + | M SAL 1 2 42 43 |
| − | M SBL 1 1 47 | + | M SBL 1 1 47 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 47 0.0319 -1.0534 | + | M SBV 1 47 0.0319 -1.0534 |
| − | S SKP 5 | + | S SKP 5 |
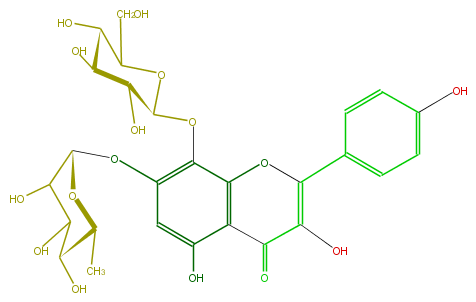
| − | ID FL5FFAGS0012 | + | ID FL5FFAGS0012 |
| − | FORMULA C27H30O16 | + | FORMULA C27H30O16 |
| − | EXACTMASS 610.153384912 | + | EXACTMASS 610.153384912 |
| − | AVERAGEMASS 610.5175 | + | AVERAGEMASS 610.5175 |
| − | SMILES C(C)(C5O)OC(C(C(O)5)O)Oc(c2)c(c(O3)c(C(=O)C(=C3c(c4)ccc(O)c4)O)c(O)2)OC(C(O)1)OC(CO)C(O)C(O)1 | + | SMILES C(C)(C5O)OC(C(C(O)5)O)Oc(c2)c(c(O3)c(C(=O)C(=C3c(c4)ccc(O)c4)O)c(O)2)OC(C(O)1)OC(CO)C(O)C(O)1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-1.6310 -0.6427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6310 -1.4676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9166 -1.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2021 -1.4676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2021 -0.6427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9166 -0.2302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5123 -1.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2268 -1.4676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2268 -0.6427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5123 -0.2302 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5123 -2.5235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1263 -0.0715 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8545 -0.4918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5827 -0.0715 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5827 0.7694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8545 1.1899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1263 0.7694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9166 -2.5344 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.3620 1.2194 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9502 -1.9937 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4845 -0.1499 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9166 0.5944 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7701 2.4415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9025 1.6204 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2139 1.3290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7105 0.7197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5517 1.5409 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3463 1.6998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3994 2.5672 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2588 2.0109 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0787 0.4355 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5883 -2.1406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8731 -1.5579 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3193 -0.8822 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3233 0.0050 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7971 -0.6844 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3764 -1.4255 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2621 -2.7532 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9693 -2.3913 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0240 -1.9801 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3620 -0.9422 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3782 2.7532 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4607 2.2234 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
20 8 1 0 0 0 0
4 3 1 0 0 0 0
1 21 1 0 0 0 0
6 22 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
25 31 1 0 0 0 0
22 26 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
37 40 1 0 0 0 0
35 21 1 0 0 0 0
36 41 1 0 0 0 0
42 43 1 0 0 0 0
28 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 42 43
M SBL 1 1 47
M SMT 1 ^CH2OH
M SBV 1 47 0.0319 -1.0534
S SKP 5
ID FL5FFAGS0012
FORMULA C27H30O16
EXACTMASS 610.153384912
AVERAGEMASS 610.5175
SMILES C(C)(C5O)OC(C(C(O)5)O)Oc(c2)c(c(O3)c(C(=O)C(=C3c(c4)ccc(O)c4)O)c(O)2)OC(C(O)1)OC(CO)C(O)C(O)1
M END