Mol:FL5FECGS0043
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 47 51 0 0 0 0 0 0 0 0999 V2000 | + | 47 51 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.0546 0.5095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0546 0.5095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0546 -0.3155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0546 -0.3155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3401 -0.7280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3401 -0.7280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3743 -0.3155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3743 -0.3155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3743 0.5095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3743 0.5095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3401 0.9221 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3401 0.9221 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0888 -0.7280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0888 -0.7280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8032 -0.3155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8032 -0.3155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8032 0.5095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8032 0.5095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0888 0.9221 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0888 0.9221 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0888 -1.5347 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0888 -1.5347 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7029 1.0808 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7029 1.0808 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4310 0.6604 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4310 0.6604 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1592 1.0808 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1592 1.0808 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1592 1.9216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1592 1.9216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4310 2.3420 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4310 2.3420 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7029 1.9216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7029 1.9216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3401 -1.5527 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3401 -1.5527 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9385 2.3715 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9385 2.3715 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8186 0.8795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8186 0.8795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4920 -0.7705 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4920 -0.7705 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1801 -2.0736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1801 -2.0736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8677 -2.4706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8677 -2.4706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6495 -1.7070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6495 -1.7070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8677 -0.9390 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8677 -0.9390 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1801 -0.5419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1801 -0.5419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3982 -1.3054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3982 -1.3054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2413 -1.3054 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2413 -1.3054 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2549 -2.8166 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2549 -2.8166 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8278 -3.1826 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8278 -3.1826 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1441 -2.0179 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1441 -2.0179 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2508 0.7772 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2508 0.7772 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5763 0.3877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5763 0.3877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7903 1.1367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7903 1.1367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5763 1.8900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5763 1.8900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2508 2.2796 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2508 2.2796 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0369 1.5306 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0369 1.5306 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8205 2.7099 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8205 2.7099 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5763 2.3760 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5763 2.3760 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6422 2.0424 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6422 2.0424 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8207 0.6245 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8207 0.6245 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4310 3.1826 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4310 3.1826 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3642 1.6932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3642 1.6932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9385 2.1087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9385 2.1087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3766 1.0964 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3766 1.0964 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6873 -0.6808 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6873 -0.6808 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7697 -1.2106 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7697 -1.2106 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 1 20 1 0 0 0 0 | + | 1 20 1 0 0 0 0 |
| − | 21 8 1 0 0 0 0 | + | 21 8 1 0 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 1 0 0 0 | + | 26 27 1 1 0 0 0 |
| − | 27 22 1 1 0 0 0 | + | 27 22 1 1 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 22 29 1 0 0 0 0 | + | 22 29 1 0 0 0 0 |
| − | 23 30 1 0 0 0 0 | + | 23 30 1 0 0 0 0 |
| − | 24 31 1 0 0 0 0 | + | 24 31 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 1 0 0 0 | + | 36 37 1 1 0 0 0 |
| − | 37 32 1 1 0 0 0 | + | 37 32 1 1 0 0 0 |
| − | 36 38 1 0 0 0 0 | + | 36 38 1 0 0 0 0 |
| − | 35 39 1 0 0 0 0 | + | 35 39 1 0 0 0 0 |
| − | 37 40 1 0 0 0 0 | + | 37 40 1 0 0 0 0 |
| − | 32 41 1 0 0 0 0 | + | 32 41 1 0 0 0 0 |
| − | 20 33 1 0 0 0 0 | + | 20 33 1 0 0 0 0 |
| − | 16 42 1 0 0 0 0 | + | 16 42 1 0 0 0 0 |
| − | 43 44 2 0 0 0 0 | + | 43 44 2 0 0 0 0 |
| − | 43 45 1 0 0 0 0 | + | 43 45 1 0 0 0 0 |
| − | 40 43 1 0 0 0 0 | + | 40 43 1 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 2 46 1 0 0 0 0 | + | 2 46 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 46 47 | + | M SAL 1 2 46 47 |
| − | M SBL 1 1 51 | + | M SBL 1 1 51 |
| − | M SMT 1 ^OCH3 | + | M SMT 1 ^OCH3 |
| − | M SBV 1 51 0.6327 0.3653 | + | M SBV 1 51 0.6327 0.3653 |
| − | S SKP 5 | + | S SKP 5 |
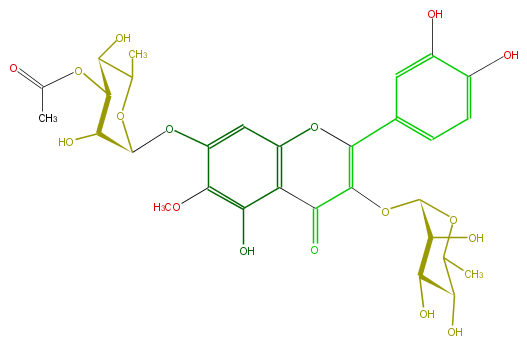
| − | ID FL5FECGS0043 | + | ID FL5FECGS0043 |
| − | FORMULA C30H34O17 | + | FORMULA C30H34O17 |
| − | EXACTMASS 666.179599662 | + | EXACTMASS 666.179599662 |
| − | AVERAGEMASS 666.58076 | + | AVERAGEMASS 666.58076 |
| − | SMILES c(O)(c5O)ccc(c5)C(O4)=C(C(=O)c(c34)c(O)c(OC)c(c3)OC(C(O)2)OC(C)C(O)C2OC(C)=O)OC(C1O)OC(C(C(O)1)O)C | + | SMILES c(O)(c5O)ccc(c5)C(O4)=C(C(=O)c(c34)c(O)c(OC)c(c3)OC(C(O)2)OC(C)C(O)C2OC(C)=O)OC(C1O)OC(C(C(O)1)O)C |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
47 51 0 0 0 0 0 0 0 0999 V2000
-1.0546 0.5095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0546 -0.3155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3401 -0.7280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3743 -0.3155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3743 0.5095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3401 0.9221 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0888 -0.7280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8032 -0.3155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8032 0.5095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0888 0.9221 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0888 -1.5347 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7029 1.0808 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4310 0.6604 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1592 1.0808 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1592 1.9216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4310 2.3420 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7029 1.9216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3401 -1.5527 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9385 2.3715 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8186 0.8795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4920 -0.7705 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1801 -2.0736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8677 -2.4706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6495 -1.7070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8677 -0.9390 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1801 -0.5419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3982 -1.3054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2413 -1.3054 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2549 -2.8166 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8278 -3.1826 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1441 -2.0179 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2508 0.7772 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5763 0.3877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7903 1.1367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5763 1.8900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2508 2.2796 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0369 1.5306 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8205 2.7099 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5763 2.3760 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6422 2.0424 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8207 0.6245 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4310 3.1826 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3642 1.6932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9385 2.1087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3766 1.0964 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6873 -0.6808 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7697 -1.2106 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
4 3 1 0 0 0 0
1 20 1 0 0 0 0
21 8 1 0 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 27 1 1 0 0 0
27 22 1 1 0 0 0
27 28 1 0 0 0 0
22 29 1 0 0 0 0
23 30 1 0 0 0 0
24 31 1 0 0 0 0
26 21 1 0 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 37 1 1 0 0 0
37 32 1 1 0 0 0
36 38 1 0 0 0 0
35 39 1 0 0 0 0
37 40 1 0 0 0 0
32 41 1 0 0 0 0
20 33 1 0 0 0 0
16 42 1 0 0 0 0
43 44 2 0 0 0 0
43 45 1 0 0 0 0
40 43 1 0 0 0 0
46 47 1 0 0 0 0
2 46 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 46 47
M SBL 1 1 51
M SMT 1 ^OCH3
M SBV 1 51 0.6327 0.3653
S SKP 5
ID FL5FECGS0043
FORMULA C30H34O17
EXACTMASS 666.179599662
AVERAGEMASS 666.58076
SMILES c(O)(c5O)ccc(c5)C(O4)=C(C(=O)c(c34)c(O)c(OC)c(c3)OC(C(O)2)OC(C)C(O)C2OC(C)=O)OC(C1O)OC(C(C(O)1)O)C
M END