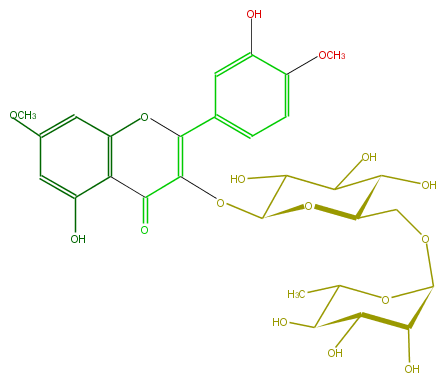
Mol:FL5FCEGL0003
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 45 49 0 0 0 0 0 0 0 0999 V2000 | + | 45 49 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.3038 1.0841 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3038 1.0841 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3038 0.2747 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3038 0.2747 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6028 -0.1300 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6028 -0.1300 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9018 0.2747 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9018 0.2747 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9018 1.0841 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9018 1.0841 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6028 1.4889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6028 1.4889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2007 -0.1300 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2007 -0.1300 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4997 0.2747 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4997 0.2747 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4997 1.0841 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4997 1.0841 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2007 1.4889 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2007 1.4889 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2007 -0.7611 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2007 -0.7611 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2011 1.4887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2011 1.4887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9156 1.0763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9156 1.0763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6299 1.4887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6299 1.4887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6299 2.3138 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6299 2.3138 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9156 2.7262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9156 2.7262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2011 2.3138 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2011 2.3138 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6028 -0.9392 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6028 -0.9392 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6756 -1.8999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6756 -1.8999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1872 -2.7460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1872 -2.7460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1266 -2.4774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1266 -2.4774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0716 -2.7460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0716 -2.7460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5681 -1.9202 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5681 -1.9202 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6207 -2.1683 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6207 -2.1683 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9390 -2.0544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9390 -2.0544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6290 0.2920 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6290 0.2920 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1406 -0.5541 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1406 -0.5541 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0799 -0.2855 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0799 -0.2855 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0250 -0.5541 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0250 -0.5541 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5136 0.2920 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5136 0.2920 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5741 0.0237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5741 0.0237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3218 -0.2221 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3218 -0.2221 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2099 0.6922 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2099 0.6922 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4136 0.1269 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4136 0.1269 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8190 -0.3698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8190 -0.3698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4057 -0.9499 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4057 -0.9499 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5688 -2.6141 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5688 -2.6141 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5346 -3.2357 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5346 -3.2357 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0730 -3.5510 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0730 -3.5510 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7277 0.2414 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7277 0.2414 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9156 3.5510 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9156 3.5510 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9382 1.5325 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9382 1.5325 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5681 2.6239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5681 2.6239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3443 2.7262 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3443 2.7262 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2446 2.2064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2446 2.2064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 20 1 0 0 0 0 | + | 19 20 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 19 25 1 0 0 0 0 | + | 19 25 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 1 0 0 0 | + | 27 28 1 1 0 0 0 |
| − | 28 29 1 1 0 0 0 | + | 28 29 1 1 0 0 0 |
| − | 30 29 1 1 0 0 0 | + | 30 29 1 1 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 26 1 0 0 0 0 | + | 31 26 1 0 0 0 0 |
| − | 31 33 1 0 0 0 0 | + | 31 33 1 0 0 0 0 |
| − | 30 34 1 0 0 0 0 | + | 30 34 1 0 0 0 0 |
| − | 29 35 1 0 0 0 0 | + | 29 35 1 0 0 0 0 |
| − | 23 36 1 0 0 0 0 | + | 23 36 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 20 37 1 0 0 0 0 | + | 20 37 1 0 0 0 0 |
| − | 21 38 1 0 0 0 0 | + | 21 38 1 0 0 0 0 |
| − | 22 39 1 0 0 0 0 | + | 22 39 1 0 0 0 0 |
| − | 32 8 1 0 0 0 0 | + | 32 8 1 0 0 0 0 |
| − | 27 32 1 0 0 0 0 | + | 27 32 1 0 0 0 0 |
| − | 26 40 1 0 0 0 0 | + | 26 40 1 0 0 0 0 |
| − | 16 41 1 0 0 0 0 | + | 16 41 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 1 42 1 0 0 0 0 | + | 1 42 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 15 44 1 0 0 0 0 | + | 15 44 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 42 43 | + | M SAL 1 2 42 43 |
| − | M SBL 1 1 47 | + | M SBL 1 1 47 |
| − | M SMT 1 ^ OCH3 | + | M SMT 1 ^ OCH3 |
| − | M SBV 1 47 0.6344 -0.4484 | + | M SBV 1 47 0.6344 -0.4484 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 44 45 | + | M SAL 2 2 44 45 |
| − | M SBL 2 1 49 | + | M SBL 2 1 49 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SBV 2 49 -0.7144 -0.4124 | + | M SBV 2 49 -0.7144 -0.4124 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FCEGL0003 | + | ID FL5FCEGL0003 |
| − | FORMULA C29H34O16 | + | FORMULA C29H34O16 |
| − | EXACTMASS 638.18468504 | + | EXACTMASS 638.18468504 |
| − | AVERAGEMASS 638.57066 | + | AVERAGEMASS 638.57066 |
| − | SMILES c(c(O)1)c(OC)cc(O4)c1C(=O)C(=C(c(c5)ccc(OC)c(O)5)4)OC(O2)C(O)C(O)C(C2COC(O3)C(C(C(O)C(C)3)O)O)O | + | SMILES c(c(O)1)c(OC)cc(O4)c1C(=O)C(=C(c(c5)ccc(OC)c(O)5)4)OC(O2)C(O)C(O)C(C2COC(O3)C(C(C(O)C(C)3)O)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
45 49 0 0 0 0 0 0 0 0999 V2000
-3.3038 1.0841 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3038 0.2747 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6028 -0.1300 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9018 0.2747 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9018 1.0841 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6028 1.4889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2007 -0.1300 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4997 0.2747 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4997 1.0841 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2007 1.4889 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2007 -0.7611 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2011 1.4887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9156 1.0763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6299 1.4887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6299 2.3138 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9156 2.7262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2011 2.3138 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6028 -0.9392 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6756 -1.8999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1872 -2.7460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1266 -2.4774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0716 -2.7460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5681 -1.9202 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6207 -2.1683 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9390 -2.0544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6290 0.2920 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1406 -0.5541 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0799 -0.2855 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0250 -0.5541 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5136 0.2920 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5741 0.0237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3218 -0.2221 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2099 0.6922 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.4136 0.1269 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8190 -0.3698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4057 -0.9499 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5688 -2.6141 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5346 -3.2357 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0730 -3.5510 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7277 0.2414 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9156 3.5510 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9382 1.5325 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5681 2.6239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3443 2.7262 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2446 2.2064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 20 1 0 0 0 0
20 21 1 1 0 0 0
21 22 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 19 1 0 0 0 0
19 25 1 0 0 0 0
26 27 1 0 0 0 0
27 28 1 1 0 0 0
28 29 1 1 0 0 0
30 29 1 1 0 0 0
30 31 1 0 0 0 0
31 26 1 0 0 0 0
31 33 1 0 0 0 0
30 34 1 0 0 0 0
29 35 1 0 0 0 0
23 36 1 0 0 0 0
35 36 1 0 0 0 0
20 37 1 0 0 0 0
21 38 1 0 0 0 0
22 39 1 0 0 0 0
32 8 1 0 0 0 0
27 32 1 0 0 0 0
26 40 1 0 0 0 0
16 41 1 0 0 0 0
42 43 1 0 0 0 0
1 42 1 0 0 0 0
44 45 1 0 0 0 0
15 44 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 42 43
M SBL 1 1 47
M SMT 1 ^ OCH3
M SBV 1 47 0.6344 -0.4484
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 44 45
M SBL 2 1 49
M SMT 2 OCH3
M SBV 2 49 -0.7144 -0.4124
S SKP 5
ID FL5FCEGL0003
FORMULA C29H34O16
EXACTMASS 638.18468504
AVERAGEMASS 638.57066
SMILES c(c(O)1)c(OC)cc(O4)c1C(=O)C(=C(c(c5)ccc(OC)c(O)5)4)OC(O2)C(O)C(O)C(C2COC(O3)C(C(C(O)C(C)3)O)O)O
M END