Mol:FL5FBCGS0002
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 35 38 0 0 0 0 0 0 0 0999 V2000 | + | 35 38 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.5750 0.4348 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5750 0.4348 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5750 -0.3749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5750 -0.3749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8738 -0.7797 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8738 -0.7797 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1725 -0.3749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1725 -0.3749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1725 0.4348 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1725 0.4348 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8738 0.8396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8738 0.8396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4712 -0.7797 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4712 -0.7797 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7700 -0.3749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7700 -0.3749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7700 0.4348 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7700 0.4348 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4712 0.8396 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4712 0.8396 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4712 -1.4719 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4712 -1.4719 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0690 0.8395 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0690 0.8395 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6455 0.4270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6455 0.4270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3602 0.8395 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3602 0.8395 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3602 1.6648 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3602 1.6648 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6455 2.0773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6455 2.0773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0690 1.6648 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0690 1.6648 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2759 0.8395 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2759 0.8395 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0150 -0.8416 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0150 -0.8416 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3151 -0.6875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3151 -0.6875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6241 -1.3785 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6241 -1.3785 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6012 -1.3624 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6012 -1.3624 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4448 -1.8664 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4448 -1.8664 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1359 -1.1754 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1359 -1.1754 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1587 -1.1915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1587 -1.1915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5326 -0.5651 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5326 -0.5651 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6379 -0.7123 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6379 -0.7123 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6955 -1.4619 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6955 -1.4619 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0747 2.0772 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0747 2.0772 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6455 2.9023 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6455 2.9023 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5405 -1.8979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5405 -1.8979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2759 -1.1626 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2759 -1.1626 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8097 -2.9023 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8097 -2.9023 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8738 -1.4501 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8738 -1.4501 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9732 -1.9701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9732 -1.9701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 20 21 1 0 0 0 0 | + | 20 21 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 25 27 1 0 0 0 0 | + | 25 27 1 0 0 0 0 |
| − | 24 28 1 0 0 0 0 | + | 24 28 1 0 0 0 0 |
| − | 21 19 1 0 0 0 0 | + | 21 19 1 0 0 0 0 |
| − | 19 8 1 0 0 0 0 | + | 19 8 1 0 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 15 29 1 0 0 0 0 | + | 15 29 1 0 0 0 0 |
| − | 16 30 1 0 0 0 0 | + | 16 30 1 0 0 0 0 |
| − | 31 32 2 0 0 0 0 | + | 31 32 2 0 0 0 0 |
| − | 31 33 1 0 0 0 0 | + | 31 33 1 0 0 0 0 |
| − | 23 31 1 0 0 0 0 | + | 23 31 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 3 34 1 0 0 0 0 | + | 3 34 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 3 31 32 33 | + | M SAL 1 3 31 32 33 |
| − | M SBL 1 1 36 | + | M SBL 1 1 36 |
| − | M SMT 1 COOH | + | M SMT 1 COOH |
| − | M SBV 1 36 -1.0957 0.0315 | + | M SBV 1 36 -1.0957 0.0315 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 34 35 | + | M SAL 2 2 34 35 |
| − | M SBL 2 1 38 | + | M SBL 2 1 38 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SBV 2 38 0.0000 0.6704 | + | M SBV 2 38 0.0000 0.6704 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FBCGS0002 | + | ID FL5FBCGS0002 |
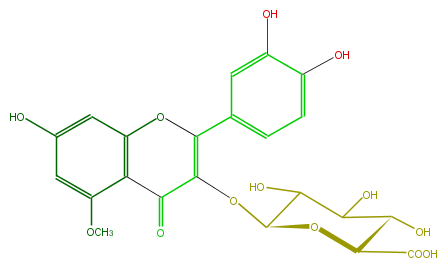
| − | FORMULA C22H20O13 | + | FORMULA C22H20O13 |
| − | EXACTMASS 492.090390726 | + | EXACTMASS 492.090390726 |
| − | AVERAGEMASS 492.3864 | + | AVERAGEMASS 492.3864 |
| − | SMILES c(c(C(O2)=C(OC(C(O)4)OC(C(O)C4O)C(O)=O)C(=O)c(c(OC)3)c2cc(c3)O)1)cc(c(c1)O)O | + | SMILES c(c(C(O2)=C(OC(C(O)4)OC(C(O)C4O)C(O)=O)C(=O)c(c(OC)3)c2cc(c3)O)1)cc(c(c1)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
35 38 0 0 0 0 0 0 0 0999 V2000
-3.5750 0.4348 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5750 -0.3749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8738 -0.7797 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1725 -0.3749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1725 0.4348 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8738 0.8396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4712 -0.7797 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7700 -0.3749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7700 0.4348 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4712 0.8396 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4712 -1.4719 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0690 0.8395 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6455 0.4270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3602 0.8395 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3602 1.6648 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6455 2.0773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0690 1.6648 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2759 0.8395 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0150 -0.8416 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3151 -0.6875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6241 -1.3785 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6012 -1.3624 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4448 -1.8664 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1359 -1.1754 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1587 -1.1915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5326 -0.5651 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6379 -0.7123 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6955 -1.4619 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0747 2.0772 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6455 2.9023 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5405 -1.8979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2759 -1.1626 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8097 -2.9023 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8738 -1.4501 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9732 -1.9701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
20 21 1 0 0 0 0
21 22 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 20 1 0 0 0 0
20 26 1 0 0 0 0
25 27 1 0 0 0 0
24 28 1 0 0 0 0
21 19 1 0 0 0 0
19 8 1 0 0 0 0
23 22 1 1 0 0 0
15 29 1 0 0 0 0
16 30 1 0 0 0 0
31 32 2 0 0 0 0
31 33 1 0 0 0 0
23 31 1 0 0 0 0
34 35 1 0 0 0 0
3 34 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 3 31 32 33
M SBL 1 1 36
M SMT 1 COOH
M SBV 1 36 -1.0957 0.0315
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 34 35
M SBL 2 1 38
M SMT 2 OCH3
M SBV 2 38 0.0000 0.6704
S SKP 5
ID FL5FBCGS0002
FORMULA C22H20O13
EXACTMASS 492.090390726
AVERAGEMASS 492.3864
SMILES c(c(C(O2)=C(OC(C(O)4)OC(C(O)C4O)C(O)=O)C(=O)c(c(OC)3)c2cc(c3)O)1)cc(c(c1)O)O
M END