Mol:FL5FADGS0005
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 35 38 0 0 0 0 0 0 0 0999 V2000 | + | 35 38 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.1631 0.2660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1631 0.2660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1632 -0.5437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1632 -0.5437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4620 -0.9484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4620 -0.9484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7611 -0.5437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7611 -0.5437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7610 0.2660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7610 0.2660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4620 0.6706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4620 0.6706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0599 -0.9484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0599 -0.9484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3589 -0.5437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3589 -0.5437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3589 0.2660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3589 0.2660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0599 0.6706 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0599 0.6706 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0599 -1.6605 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0599 -1.6605 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3419 0.6705 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3419 0.6705 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0564 0.2580 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0564 0.2580 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7711 0.6705 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7711 0.6705 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7710 1.4955 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7710 1.4955 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0564 1.9081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0564 1.9081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3419 1.4955 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3419 1.4955 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8639 0.6704 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8639 0.6704 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4122 -1.0360 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4122 -1.0360 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6088 -1.2324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6088 -1.2324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8743 -1.4998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8743 -1.4998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5881 -1.8184 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5881 -1.8184 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0292 -2.4689 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0292 -2.4689 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7638 -2.2016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7638 -2.2016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0500 -1.8830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0500 -1.8830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9440 -0.8808 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9440 -0.8808 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8509 -1.7416 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8509 -1.7416 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0974 -2.6161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0974 -2.6161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4952 1.9136 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4952 1.9136 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4620 -1.6555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4620 -1.6555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0564 2.7010 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0564 2.7010 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6127 3.8317 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6127 3.8317 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9166 -3.0126 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9166 -3.0126 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8639 -2.6678 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8639 -2.6678 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7721 -3.8317 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7721 -3.8317 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 20 21 1 0 0 0 0 | + | 20 21 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 25 27 1 0 0 0 0 | + | 25 27 1 0 0 0 0 |
| − | 24 28 1 0 0 0 0 | + | 24 28 1 0 0 0 0 |
| − | 21 19 1 0 0 0 0 | + | 21 19 1 0 0 0 0 |
| − | 19 8 1 0 0 0 0 | + | 19 8 1 0 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 15 29 1 0 0 0 0 | + | 15 29 1 0 0 0 0 |
| − | 3 30 1 0 0 0 0 | + | 3 30 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 16 31 1 0 0 0 0 | + | 16 31 1 0 0 0 0 |
| − | 33 34 2 0 0 0 0 | + | 33 34 2 0 0 0 0 |
| − | 33 35 1 0 0 0 0 | + | 33 35 1 0 0 0 0 |
| − | 23 33 1 0 0 0 0 | + | 23 33 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 31 32 | + | M SAL 1 2 31 32 |
| − | M SBL 1 1 35 | + | M SBL 1 1 35 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 35 0.0000 -0.7929 | + | M SBV 1 35 0.0000 -0.7929 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 3 33 34 35 | + | M SAL 2 3 33 34 35 |
| − | M SBL 2 1 38 | + | M SBL 2 1 38 |
| − | M SMT 2 COOH | + | M SMT 2 COOH |
| − | M SBV 2 38 -0.8873 0.5438 | + | M SBV 2 38 -0.8873 0.5438 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FADGS0005 | + | ID FL5FADGS0005 |
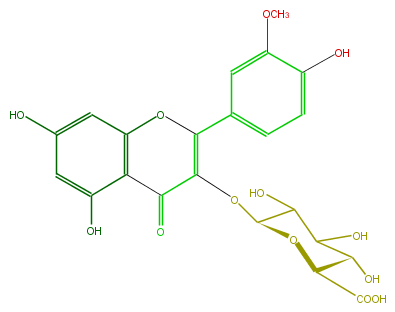
| − | FORMULA C22H20O13 | + | FORMULA C22H20O13 |
| − | EXACTMASS 492.090390726 | + | EXACTMASS 492.090390726 |
| − | AVERAGEMASS 492.3864 | + | AVERAGEMASS 492.3864 |
| − | SMILES c(C(=O)2)(c1O)c(OC(c(c4)cc(OC)c(O)c4)=C2OC(C(O)3)OC(C(O)C3O)C(O)=O)cc(c1)O | + | SMILES c(C(=O)2)(c1O)c(OC(c(c4)cc(OC)c(O)c4)=C2OC(C(O)3)OC(C(O)C3O)C(O)=O)cc(c1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
35 38 0 0 0 0 0 0 0 0999 V2000
-3.1631 0.2660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1632 -0.5437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4620 -0.9484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7611 -0.5437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7610 0.2660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4620 0.6706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0599 -0.9484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3589 -0.5437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3589 0.2660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0599 0.6706 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0599 -1.6605 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3419 0.6705 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0564 0.2580 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7711 0.6705 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7710 1.4955 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0564 1.9081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3419 1.4955 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8639 0.6704 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4122 -1.0360 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6088 -1.2324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8743 -1.4998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5881 -1.8184 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0292 -2.4689 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7638 -2.2016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0500 -1.8830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9440 -0.8808 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8509 -1.7416 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0974 -2.6161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4952 1.9136 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4620 -1.6555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0564 2.7010 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6127 3.8317 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9166 -3.0126 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8639 -2.6678 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7721 -3.8317 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
20 21 1 0 0 0 0
21 22 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 20 1 0 0 0 0
20 26 1 0 0 0 0
25 27 1 0 0 0 0
24 28 1 0 0 0 0
21 19 1 0 0 0 0
19 8 1 0 0 0 0
23 22 1 1 0 0 0
15 29 1 0 0 0 0
3 30 1 0 0 0 0
31 32 1 0 0 0 0
16 31 1 0 0 0 0
33 34 2 0 0 0 0
33 35 1 0 0 0 0
23 33 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 31 32
M SBL 1 1 35
M SMT 1 OCH3
M SBV 1 35 0.0000 -0.7929
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 3 33 34 35
M SBL 2 1 38
M SMT 2 COOH
M SBV 2 38 -0.8873 0.5438
S SKP 5
ID FL5FADGS0005
FORMULA C22H20O13
EXACTMASS 492.090390726
AVERAGEMASS 492.3864
SMILES c(C(=O)2)(c1O)c(OC(c(c4)cc(OC)c(O)c4)=C2OC(C(O)3)OC(C(O)C3O)C(O)=O)cc(c1)O
M END