Mol:FL5FADGA0017
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 55 60 0 0 0 0 0 0 0 0999 V2000 | + | 55 60 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.6473 1.2794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6473 1.2794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6473 0.6371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6473 0.6371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0910 0.3159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0910 0.3159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4653 0.6371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4653 0.6371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4653 1.2794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4653 1.2794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0910 1.6006 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0910 1.6006 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0216 0.3159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0216 0.3159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5779 0.6371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5779 0.6371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5779 1.2794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5779 1.2794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0216 1.6006 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0216 1.6006 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0216 -0.1850 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0216 -0.1850 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1340 1.6005 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1340 1.6005 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7009 1.2731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7009 1.2731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2679 1.6005 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2679 1.6005 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2679 2.2552 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2679 2.2552 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7009 2.5825 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7009 2.5825 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1340 2.2552 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1340 2.2552 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0910 -0.3262 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0910 -0.3262 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2034 1.6005 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2034 1.6005 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1339 2.7552 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1339 2.7552 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9720 0.2482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9720 0.2482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7606 -0.7790 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.7606 -0.7790 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.0892 -0.3914 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.0892 -0.3914 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.3023 -1.1368 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3023 -1.1368 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0892 -1.8868 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.0892 -1.8868 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.7606 -2.2745 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.7606 -2.2745 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.5476 -1.5290 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.5476 -1.5290 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.5677 -0.0590 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5677 -0.0590 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1592 -1.5824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1592 -1.5824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9891 -2.9022 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9891 -2.9022 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6378 -0.8809 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 3.6378 -0.8809 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 4.3632 -0.8809 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 4.3632 -0.8809 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.8418 -0.3765 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 3.8418 -0.3765 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 3.6636 0.3309 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6636 0.3309 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9381 0.3309 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.9381 0.3309 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.4595 -0.1735 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.4595 -0.1735 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 4.1267 0.2116 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1267 0.2116 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0786 -1.3218 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0786 -1.3218 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5148 -1.4466 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5148 -1.4466 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5580 -0.3765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5580 -0.3765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8442 1.4444 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -3.8442 1.4444 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -3.3286 0.7638 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.3286 0.7638 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.5861 1.0525 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.5861 1.0525 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.8696 1.0603 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.8696 1.0603 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.3903 1.5810 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3903 1.5810 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0398 1.2382 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -3.0398 1.2382 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -4.5580 1.0323 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5580 1.0323 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1324 0.7247 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1324 0.7247 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1607 0.3384 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1607 0.3384 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9843 3.1586 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9843 3.1586 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4257 4.0559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4257 4.0559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4446 -2.4191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4446 -2.4191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9144 -3.0511 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9144 -3.0511 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2449 2.1633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2449 2.1633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1986 2.4640 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1986 2.4640 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 8 21 1 0 0 0 0 | + | 8 21 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 23 21 1 0 0 0 0 | + | 23 21 1 0 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 1 0 0 0 | + | 35 36 1 1 0 0 0 |
| − | 36 31 1 1 0 0 0 | + | 36 31 1 1 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 31 38 1 0 0 0 0 | + | 31 38 1 0 0 0 0 |
| − | 32 39 1 0 0 0 0 | + | 32 39 1 0 0 0 0 |
| − | 33 40 1 0 0 0 0 | + | 33 40 1 0 0 0 0 |
| − | 35 28 1 0 0 0 0 | + | 35 28 1 0 0 0 0 |
| − | 41 42 1 1 0 0 0 | + | 41 42 1 1 0 0 0 |
| − | 42 43 1 1 0 0 0 | + | 42 43 1 1 0 0 0 |
| − | 44 43 1 1 0 0 0 | + | 44 43 1 1 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 46 41 1 0 0 0 0 | + | 46 41 1 0 0 0 0 |
| − | 41 47 1 0 0 0 0 | + | 41 47 1 0 0 0 0 |
| − | 42 48 1 0 0 0 0 | + | 42 48 1 0 0 0 0 |
| − | 43 49 1 0 0 0 0 | + | 43 49 1 0 0 0 0 |
| − | 44 19 1 0 0 0 0 | + | 44 19 1 0 0 0 0 |
| − | 16 50 1 0 0 0 0 | + | 16 50 1 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 25 52 1 0 0 0 0 | + | 25 52 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 46 54 1 0 0 0 0 | + | 46 54 1 0 0 0 0 |
| − | 54 55 1 0 0 0 0 | + | 54 55 1 0 0 0 0 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 54 55 | + | M SAL 3 2 54 55 |
| − | M SBL 3 1 59 | + | M SBL 3 1 59 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SVB 3 59 -3.2449 2.1633 | + | M SVB 3 59 -3.2449 2.1633 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 52 53 | + | M SAL 2 2 52 53 |
| − | M SBL 2 1 57 | + | M SBL 2 1 57 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 57 2.5633 -2.5753 | + | M SVB 2 57 2.5633 -2.5753 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 50 51 | + | M SAL 1 2 50 51 |
| − | M SBL 1 1 55 | + | M SBL 1 1 55 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 55 2.9843 3.1586 | + | M SVB 1 55 2.9843 3.1586 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FADGA0017 | + | ID FL5FADGA0017 |
| − | KNApSAcK_ID C00005576 | + | KNApSAcK_ID C00005576 |
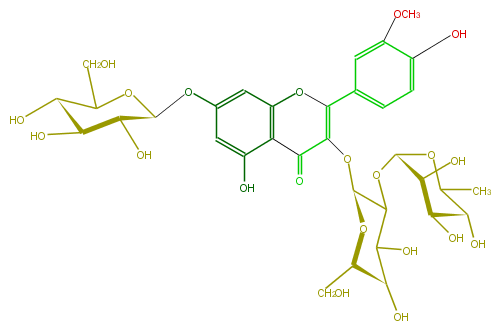
| − | NAME Isorhamnetin 3-rhamnosyl-(1->2)-galactoside-7-glucoside | + | NAME Isorhamnetin 3-rhamnosyl-(1->2)-galactoside-7-glucoside |
| − | CAS_RN 128988-18-1 | + | CAS_RN 128988-18-1 |
| − | FORMULA C34H42O21 | + | FORMULA C34H42O21 |
| − | EXACTMASS 786.2218584059999 | + | EXACTMASS 786.2218584059999 |
| − | AVERAGEMASS 786.68468 | + | AVERAGEMASS 786.68468 |
| − | SMILES O(C)c(c1O)cc(C(=C4O[C@@H](C5O[C@@H](O6)[C@@H](O)[C@H](O)[C@@H](C6C)O)O[C@H](CO)[C@@H](C5O)O)Oc(c3C4=O)cc(cc3O)O[C@@H]([C@@H](O)2)OC([C@@H]([C@H](O)2)O)CO)cc1 | + | SMILES O(C)c(c1O)cc(C(=C4O[C@@H](C5O[C@@H](O6)[C@@H](O)[C@H](O)[C@@H](C6C)O)O[C@H](CO)[C@@H](C5O)O)Oc(c3C4=O)cc(cc3O)O[C@@H]([C@@H](O)2)OC([C@@H]([C@H](O)2)O)CO)cc1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
55 60 0 0 0 0 0 0 0 0999 V2000
-0.6473 1.2794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6473 0.6371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0910 0.3159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4653 0.6371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4653 1.2794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0910 1.6006 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0216 0.3159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5779 0.6371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5779 1.2794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0216 1.6006 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0216 -0.1850 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1340 1.6005 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7009 1.2731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2679 1.6005 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2679 2.2552 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7009 2.5825 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1340 2.2552 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0910 -0.3262 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2034 1.6005 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1339 2.7552 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9720 0.2482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7606 -0.7790 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.0892 -0.3914 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.3023 -1.1368 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0892 -1.8868 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.7606 -2.2745 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.5476 -1.5290 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.5677 -0.0590 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1592 -1.5824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9891 -2.9022 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6378 -0.8809 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
4.3632 -0.8809 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.8418 -0.3765 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
3.6636 0.3309 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9381 0.3309 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.4595 -0.1735 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
4.1267 0.2116 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0786 -1.3218 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5148 -1.4466 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5580 -0.3765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8442 1.4444 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-3.3286 0.7638 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.5861 1.0525 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.8696 1.0603 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.3903 1.5810 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0398 1.2382 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-4.5580 1.0323 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1324 0.7247 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1607 0.3384 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9843 3.1586 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4257 4.0559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4446 -2.4191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9144 -3.0511 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2449 2.1633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1986 2.4640 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
15 20 1 0 0 0 0
8 21 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
23 21 1 0 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 36 1 1 0 0 0
36 31 1 1 0 0 0
36 37 1 0 0 0 0
31 38 1 0 0 0 0
32 39 1 0 0 0 0
33 40 1 0 0 0 0
35 28 1 0 0 0 0
41 42 1 1 0 0 0
42 43 1 1 0 0 0
44 43 1 1 0 0 0
44 45 1 0 0 0 0
45 46 1 0 0 0 0
46 41 1 0 0 0 0
41 47 1 0 0 0 0
42 48 1 0 0 0 0
43 49 1 0 0 0 0
44 19 1 0 0 0 0
16 50 1 0 0 0 0
50 51 1 0 0 0 0
25 52 1 0 0 0 0
52 53 1 0 0 0 0
46 54 1 0 0 0 0
54 55 1 0 0 0 0
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 54 55
M SBL 3 1 59
M SMT 3 CH2OH
M SVB 3 59 -3.2449 2.1633
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 52 53
M SBL 2 1 57
M SMT 2 CH2OH
M SVB 2 57 2.5633 -2.5753
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 50 51
M SBL 1 1 55
M SMT 1 OCH3
M SVB 1 55 2.9843 3.1586
S SKP 8
ID FL5FADGA0017
KNApSAcK_ID C00005576
NAME Isorhamnetin 3-rhamnosyl-(1->2)-galactoside-7-glucoside
CAS_RN 128988-18-1
FORMULA C34H42O21
EXACTMASS 786.2218584059999
AVERAGEMASS 786.68468
SMILES O(C)c(c1O)cc(C(=C4O[C@@H](C5O[C@@H](O6)[C@@H](O)[C@H](O)[C@@H](C6C)O)O[C@H](CO)[C@@H](C5O)O)Oc(c3C4=O)cc(cc3O)O[C@@H]([C@@H](O)2)OC([C@@H]([C@H](O)2)O)CO)cc1
M END