Mol:FL5FADGA0004
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 45 49 0 0 0 0 0 0 0 0999 V2000 | + | 45 49 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.8762 0.9823 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8762 0.9823 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8762 0.3400 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8762 0.3400 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3199 0.0188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3199 0.0188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7636 0.3400 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7636 0.3400 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7636 0.9823 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7636 0.9823 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3199 1.3035 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3199 1.3035 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2073 0.0188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2073 0.0188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6510 0.3400 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6510 0.3400 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6510 0.9823 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6510 0.9823 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2073 1.3035 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2073 1.3035 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2073 -0.4820 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2073 -0.4820 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0949 1.3034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0949 1.3034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4721 0.9761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4721 0.9761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0391 1.3034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0391 1.3034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0391 1.9581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0391 1.9581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4721 2.2854 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4721 2.2854 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0949 1.9581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0949 1.9581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4323 1.3034 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4323 1.3034 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2568 -0.0489 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2568 -0.0489 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3199 -0.6233 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3199 -0.6233 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9051 2.4581 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9051 2.4581 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1338 -1.1667 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 0.1338 -1.1667 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -0.4490 -0.8302 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.4490 -0.8302 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.2641 -1.4773 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2641 -1.4773 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4490 -2.1282 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.4490 -2.1282 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.1338 -2.4648 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.1338 -2.4648 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.0511 -1.8177 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -0.0511 -1.8177 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.4104 -2.0841 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4104 -2.0841 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8402 -2.2755 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8402 -2.2755 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2011 -1.1845 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 1.2011 -1.1845 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 1.6435 -1.7685 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.6435 -1.7685 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.2805 -1.5207 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.2805 -1.5207 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.8953 -1.5141 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.8953 -1.5141 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.4486 -1.0673 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.4486 -1.0673 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.8912 -1.3615 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8912 -1.3615 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6456 -2.1335 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6456 -2.1335 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8865 -1.6462 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8865 -1.6462 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3990 -0.9015 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3990 -0.9015 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6088 -2.1659 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6088 -2.1659 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7555 2.8616 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7555 2.8616 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1968 3.7589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1968 3.7589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2055 -0.4138 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2055 -0.4138 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0181 0.5685 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0181 0.5685 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8062 -2.1512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8062 -2.1512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5206 -2.5636 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5206 -2.5636 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 8 19 1 0 0 0 0 | + | 8 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 21 15 1 0 0 0 0 | + | 21 15 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 26 29 1 0 0 0 0 | + | 26 29 1 0 0 0 0 |
| − | 23 19 1 0 0 0 0 | + | 23 19 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 32 36 1 0 0 0 0 | + | 32 36 1 0 0 0 0 |
| − | 33 37 1 0 0 0 0 | + | 33 37 1 0 0 0 0 |
| − | 22 38 1 0 0 0 0 | + | 22 38 1 0 0 0 0 |
| − | 31 39 1 0 0 0 0 | + | 31 39 1 0 0 0 0 |
| − | 30 28 1 0 0 0 0 | + | 30 28 1 0 0 0 0 |
| − | 16 40 1 0 0 0 0 | + | 16 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 34 42 1 0 0 0 0 | + | 34 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 25 44 1 0 0 0 0 | + | 25 44 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 44 45 | + | M SAL 3 2 44 45 |
| − | M SBL 3 1 48 | + | M SBL 3 1 48 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SVB 3 48 -0.6078 -2.4491 | + | M SVB 3 48 -0.6078 -2.4491 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 42 43 | + | M SAL 2 2 42 43 |
| − | M SBL 2 1 46 | + | M SBL 2 1 46 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 46 3.2592 -1.3807 | + | M SVB 2 46 3.2592 -1.3807 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 44 | + | M SBL 1 1 44 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 44 0.7555 2.8616 | + | M SVB 1 44 0.7555 2.8616 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FADGA0004 | + | ID FL5FADGA0004 |
| − | KNApSAcK_ID C00005535 | + | KNApSAcK_ID C00005535 |
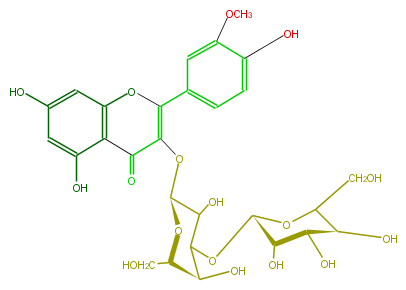
| − | NAME Isorhamnetin 3-glucosyl-(1->3)-galactoside | + | NAME Isorhamnetin 3-glucosyl-(1->3)-galactoside |
| − | CAS_RN 139955-71-8 | + | CAS_RN 139955-71-8 |
| − | FORMULA C28H32O17 | + | FORMULA C28H32O17 |
| − | EXACTMASS 640.163949598 | + | EXACTMASS 640.163949598 |
| − | AVERAGEMASS 640.54348 | + | AVERAGEMASS 640.54348 |
| − | SMILES C(O[C@H]([C@@H]5O)OC([C@H]([C@@H](O)5)O)CO)([C@H]1O)C(O)[C@H](OC(=C2c(c4)ccc(c4OC)O)C(c(c3O)c(cc(O)c3)O2)=O)O[C@H](CO)1 | + | SMILES C(O[C@H]([C@@H]5O)OC([C@H]([C@@H](O)5)O)CO)([C@H]1O)C(O)[C@H](OC(=C2c(c4)ccc(c4OC)O)C(c(c3O)c(cc(O)c3)O2)=O)O[C@H](CO)1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
45 49 0 0 0 0 0 0 0 0999 V2000
-2.8762 0.9823 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8762 0.3400 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3199 0.0188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7636 0.3400 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7636 0.9823 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3199 1.3035 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2073 0.0188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6510 0.3400 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6510 0.9823 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2073 1.3035 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2073 -0.4820 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0949 1.3034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4721 0.9761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0391 1.3034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0391 1.9581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4721 2.2854 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0949 1.9581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4323 1.3034 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2568 -0.0489 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3199 -0.6233 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9051 2.4581 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1338 -1.1667 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-0.4490 -0.8302 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.2641 -1.4773 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4490 -2.1282 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.1338 -2.4648 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.0511 -1.8177 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.4104 -2.0841 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8402 -2.2755 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2011 -1.1845 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
1.6435 -1.7685 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.2805 -1.5207 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.8953 -1.5141 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.4486 -1.0673 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.8912 -1.3615 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6456 -2.1335 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8865 -1.6462 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3990 -0.9015 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6088 -2.1659 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7555 2.8616 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1968 3.7589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2055 -0.4138 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0181 0.5685 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8062 -2.1512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5206 -2.5636 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
8 19 1 0 0 0 0
3 20 1 0 0 0 0
21 15 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
27 28 1 0 0 0 0
26 29 1 0 0 0 0
23 19 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
32 36 1 0 0 0 0
33 37 1 0 0 0 0
22 38 1 0 0 0 0
31 39 1 0 0 0 0
30 28 1 0 0 0 0
16 40 1 0 0 0 0
40 41 1 0 0 0 0
34 42 1 0 0 0 0
42 43 1 0 0 0 0
25 44 1 0 0 0 0
44 45 1 0 0 0 0
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 44 45
M SBL 3 1 48
M SMT 3 CH2OH
M SVB 3 48 -0.6078 -2.4491
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 42 43
M SBL 2 1 46
M SMT 2 CH2OH
M SVB 2 46 3.2592 -1.3807
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 44
M SMT 1 OCH3
M SVB 1 44 0.7555 2.8616
S SKP 8
ID FL5FADGA0004
KNApSAcK_ID C00005535
NAME Isorhamnetin 3-glucosyl-(1->3)-galactoside
CAS_RN 139955-71-8
FORMULA C28H32O17
EXACTMASS 640.163949598
AVERAGEMASS 640.54348
SMILES C(O[C@H]([C@@H]5O)OC([C@H]([C@@H](O)5)O)CO)([C@H]1O)C(O)[C@H](OC(=C2c(c4)ccc(c4OC)O)C(c(c3O)c(cc(O)c3)O2)=O)O[C@H](CO)1
M END