Mol:FL5FACGSS002
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 38 41 0 0 0 0 0 0 0 0999 V2000 | + | 38 41 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.1053 0.1949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1053 0.1949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1053 -0.6149 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1053 -0.6149 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4041 -1.0198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4041 -1.0198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7030 -0.6149 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7030 -0.6149 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7029 0.1949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7029 0.1949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4041 0.5997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4041 0.5997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0017 -1.0198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0017 -1.0198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3006 -0.6149 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3006 -0.6149 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3005 0.1949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3005 0.1949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0018 0.5997 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0018 0.5997 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0016 -1.6509 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0016 -1.6509 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4004 0.5995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4004 0.5995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1150 0.1869 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1150 0.1869 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8298 0.5994 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8298 0.5994 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8297 1.4246 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8297 1.4246 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1150 1.8373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1150 1.8373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4004 1.4246 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4004 1.4246 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4439 -1.0533 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4439 -1.0533 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5635 -1.4038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5635 -1.4038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8288 -1.6711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8288 -1.6711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5428 -1.9897 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5428 -1.9897 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9840 -2.6405 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9840 -2.6405 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7187 -2.3731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7187 -2.3731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0047 -2.0545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0047 -2.0545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1027 -1.0014 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1027 -1.0014 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6393 -1.8910 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6393 -1.8910 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9685 -2.7655 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9685 -2.7655 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4796 1.7998 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4796 1.7998 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4041 -1.6723 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4041 -1.6723 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7487 0.5663 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7487 0.5663 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1151 2.6623 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1151 2.6623 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1066 3.9825 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1066 3.9825 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1066 3.3474 0.0000 S 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1066 3.3474 0.0000 S 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7310 3.3474 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7310 3.3474 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5339 3.3474 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5339 3.3474 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8012 -3.1632 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8012 -3.1632 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7487 -2.8184 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7487 -2.8184 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6567 -3.9825 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6567 -3.9825 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 19 20 1 0 0 0 0 | + | 19 20 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 19 25 1 0 0 0 0 | + | 19 25 1 0 0 0 0 |
| − | 24 26 1 0 0 0 0 | + | 24 26 1 0 0 0 0 |
| − | 23 27 1 0 0 0 0 | + | 23 27 1 0 0 0 0 |
| − | 20 18 1 0 0 0 0 | + | 20 18 1 0 0 0 0 |
| − | 18 8 1 0 0 0 0 | + | 18 8 1 0 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 15 28 1 0 0 0 0 | + | 15 28 1 0 0 0 0 |
| − | 3 29 1 0 0 0 0 | + | 3 29 1 0 0 0 0 |
| − | 1 30 1 0 0 0 0 | + | 1 30 1 0 0 0 0 |
| − | 16 31 1 0 0 0 0 | + | 16 31 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 2 0 0 0 0 | + | 33 34 2 0 0 0 0 |
| − | 33 35 2 0 0 0 0 | + | 33 35 2 0 0 0 0 |
| − | 31 33 1 0 0 0 0 | + | 31 33 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 36 38 1 0 0 0 0 | + | 36 38 1 0 0 0 0 |
| − | 22 36 1 0 0 0 0 | + | 22 36 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 3 36 37 38 | + | M SAL 1 3 36 37 38 |
| − | M SBL 1 1 41 | + | M SBL 1 1 41 |
| − | M SMT 1 COOH | + | M SMT 1 COOH |
| − | M SBV 1 41 -0.8172 0.5228 | + | M SBV 1 41 -0.8172 0.5228 |
| − | S SKP 5 | + | S SKP 5 |
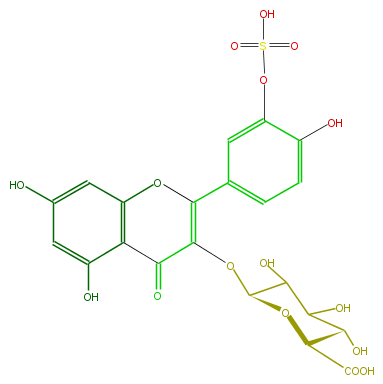
| − | ID FL5FACGSS002 | + | ID FL5FACGSS002 |
| − | FORMULA C21H18O16S | + | FORMULA C21H18O16S |
| − | EXACTMASS 558.031555218 | + | EXACTMASS 558.031555218 |
| − | AVERAGEMASS 558.42402 | + | AVERAGEMASS 558.42402 |
| − | SMILES C(C(C(O)=O)1)(O)C(C(C(OC(=C3c(c4)cc(c(c4)O)OS(O)(=O)=O)C(=O)c(c(O3)2)c(cc(O)c2)O)O1)O)O | + | SMILES C(C(C(O)=O)1)(O)C(C(C(OC(=C3c(c4)cc(c(c4)O)OS(O)(=O)=O)C(=O)c(c(O3)2)c(cc(O)c2)O)O1)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
38 41 0 0 0 0 0 0 0 0999 V2000
-3.1053 0.1949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1053 -0.6149 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4041 -1.0198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7030 -0.6149 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7029 0.1949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4041 0.5997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0017 -1.0198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3006 -0.6149 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3005 0.1949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0018 0.5997 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0016 -1.6509 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4004 0.5995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1150 0.1869 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8298 0.5994 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8297 1.4246 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1150 1.8373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4004 1.4246 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4439 -1.0533 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5635 -1.4038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8288 -1.6711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5428 -1.9897 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9840 -2.6405 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7187 -2.3731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0047 -2.0545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1027 -1.0014 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6393 -1.8910 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9685 -2.7655 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4796 1.7998 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4041 -1.6723 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7487 0.5663 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1151 2.6623 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1066 3.9825 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1066 3.3474 0.0000 S 0 0 0 0 0 0 0 0 0 0 0 0
1.7310 3.3474 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5339 3.3474 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8012 -3.1632 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7487 -2.8184 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6567 -3.9825 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
19 20 1 0 0 0 0
20 21 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 19 1 0 0 0 0
19 25 1 0 0 0 0
24 26 1 0 0 0 0
23 27 1 0 0 0 0
20 18 1 0 0 0 0
18 8 1 0 0 0 0
22 21 1 1 0 0 0
15 28 1 0 0 0 0
3 29 1 0 0 0 0
1 30 1 0 0 0 0
16 31 1 0 0 0 0
32 33 1 0 0 0 0
33 34 2 0 0 0 0
33 35 2 0 0 0 0
31 33 1 0 0 0 0
36 37 2 0 0 0 0
36 38 1 0 0 0 0
22 36 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 3 36 37 38
M SBL 1 1 41
M SMT 1 COOH
M SBV 1 41 -0.8172 0.5228
S SKP 5
ID FL5FACGSS002
FORMULA C21H18O16S
EXACTMASS 558.031555218
AVERAGEMASS 558.42402
SMILES C(C(C(O)=O)1)(O)C(C(C(OC(=C3c(c4)cc(c(c4)O)OS(O)(=O)=O)C(=O)c(c(O3)2)c(cc(O)c2)O)O1)O)O
M END