Mol:FL5FACGS0104
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 56 61 0 0 0 0 0 0 0 0999 V2000 | + | 56 61 0 0 0 0 0 0 0 0999 V2000 |
| − | 4.2899 1.3375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2899 1.3375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5754 1.7500 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5754 1.7500 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5754 2.5750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5754 2.5750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2899 2.9875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2899 2.9875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0044 2.5750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0044 2.5750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0044 1.7500 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0044 1.7500 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8610 1.3375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8610 1.3375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1465 1.7500 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1465 1.7500 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4320 1.3375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4320 1.3375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4320 0.5125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4320 0.5125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1465 0.1000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1465 0.1000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8610 0.5125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8610 0.5125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7175 1.7500 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7175 1.7500 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0031 1.3375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0031 1.3375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0031 0.5125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0031 0.5125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7175 0.1000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7175 0.1000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1465 -0.5608 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1465 -0.5608 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7114 1.7500 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7114 1.7500 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4599 0.0670 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4599 0.0670 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7175 -0.6267 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7175 -0.6267 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.5963 2.9167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.5963 2.9167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2899 3.7133 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2899 3.7133 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0031 -1.7616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0031 -1.7616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8087 -1.9407 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8087 -1.9407 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1943 -1.2110 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1943 -1.2110 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8979 -0.7797 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8979 -0.7797 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0923 -0.6005 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0923 -0.6005 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7066 -1.3301 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7066 -1.3301 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1688 -1.0498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1688 -1.0498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6403 -1.2161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6403 -1.2161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0031 -2.4631 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0031 -2.4631 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3145 -2.0962 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3145 -2.0962 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5500 -1.7700 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5500 -1.7700 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1999 -2.9425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1999 -2.9425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6057 -3.1216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6057 -3.1216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9913 -2.3919 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9913 -2.3919 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6949 -1.9606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6949 -1.9606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8893 -1.7814 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8893 -1.7814 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5036 -2.5111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5036 -2.5111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0342 -2.2307 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0342 -2.2307 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1999 -3.7133 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1999 -3.7133 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1115 -3.2771 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1115 -3.2771 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3469 -2.9509 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3469 -2.9509 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6507 -2.2923 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6507 -2.2923 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0627 -1.5788 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0627 -1.5788 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6507 -0.8653 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6507 -0.8653 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8865 -1.5788 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8865 -1.5788 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2985 -2.2923 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2985 -2.2923 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1224 -2.2923 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1224 -2.2923 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5349 -1.5778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5349 -1.5778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3599 -1.5778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3599 -1.5778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7724 -2.2923 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7724 -2.2923 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3599 -3.0068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3599 -3.0068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5349 -3.0068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5349 -3.0068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.5963 -2.2923 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.5963 -2.2923 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7718 -0.8643 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7718 -0.8643 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 2 7 1 0 0 0 0 | + | 2 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 11 1 0 0 0 0 | + | 10 11 1 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 7 2 0 0 0 0 | + | 12 7 2 0 0 0 0 |
| − | 9 13 1 0 0 0 0 | + | 9 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 10 1 0 0 0 0 | + | 16 10 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 11 17 2 0 0 0 0 | + | 11 17 2 0 0 0 0 |
| − | 12 19 1 0 0 0 0 | + | 12 19 1 0 0 0 0 |
| − | 16 20 1 0 0 0 0 | + | 16 20 1 0 0 0 0 |
| − | 5 21 1 0 0 0 0 | + | 5 21 1 0 0 0 0 |
| − | 4 22 1 0 0 0 0 | + | 4 22 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 23 31 1 0 0 0 0 | + | 23 31 1 0 0 0 0 |
| − | 24 32 1 0 0 0 0 | + | 24 32 1 0 0 0 0 |
| − | 25 33 1 0 0 0 0 | + | 25 33 1 0 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 35 36 1 1 0 0 0 | + | 35 36 1 1 0 0 0 |
| − | 37 36 1 1 0 0 0 | + | 37 36 1 1 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 39 34 1 0 0 0 0 | + | 39 34 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 34 41 1 0 0 0 0 | + | 34 41 1 0 0 0 0 |
| − | 35 42 1 0 0 0 0 | + | 35 42 1 0 0 0 0 |
| − | 36 43 1 0 0 0 0 | + | 36 43 1 0 0 0 0 |
| − | 37 30 1 0 0 0 0 | + | 37 30 1 0 0 0 0 |
| − | 26 19 1 0 0 0 0 | + | 26 19 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 46 2 0 0 0 0 | + | 45 46 2 0 0 0 0 |
| − | 45 47 1 0 0 0 0 | + | 45 47 1 0 0 0 0 |
| − | 47 48 2 0 0 0 0 | + | 47 48 2 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 49 50 2 0 0 0 0 | + | 49 50 2 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 51 52 2 0 0 0 0 | + | 51 52 2 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 53 54 2 0 0 0 0 | + | 53 54 2 0 0 0 0 |
| − | 54 49 1 0 0 0 0 | + | 54 49 1 0 0 0 0 |
| − | 52 55 1 0 0 0 0 | + | 52 55 1 0 0 0 0 |
| − | 51 56 1 0 0 0 0 | + | 51 56 1 0 0 0 0 |
| − | 44 40 1 0 0 0 0 | + | 44 40 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FACGS0104 | + | ID FL5FACGS0104 |
| − | KNApSAcK_ID C00013876 | + | KNApSAcK_ID C00013876 |
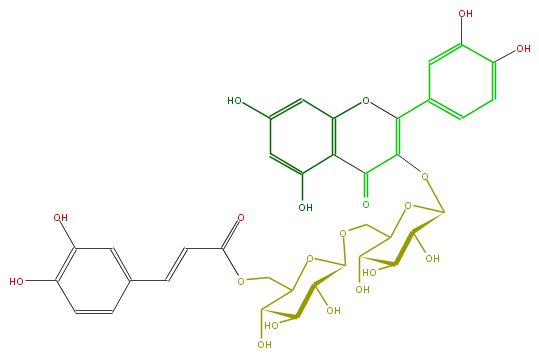
| − | NAME Madreselvin B;Quercetin 3-(6'''-caffeoylgentiobioside);2-(3,4-Dihydroxyphenyl)-3-[[6-O-[6-O-[(2E)-3-(3,4-dihydroxyphenyl)-1-oxo-2-propenyl]-beta-D-glucopyranosyl]-beta-D-glucopyranosyl]oxy]-5,7-dihydroxy-4H-1-benzopyran-4-one | + | NAME Madreselvin B;Quercetin 3-(6'''-caffeoylgentiobioside);2-(3,4-Dihydroxyphenyl)-3-[[6-O-[6-O-[(2E)-3-(3,4-dihydroxyphenyl)-1-oxo-2-propenyl]-beta-D-glucopyranosyl]-beta-D-glucopyranosyl]oxy]-5,7-dihydroxy-4H-1-benzopyran-4-one |
| − | CAS_RN 188682-90-8 | + | CAS_RN 188682-90-8 |
| − | FORMULA C36H36O20 | + | FORMULA C36H36O20 |
| − | EXACTMASS 788.179993592 | + | EXACTMASS 788.179993592 |
| − | AVERAGEMASS 788.65904 | + | AVERAGEMASS 788.65904 |
| − | SMILES C(C4O)(COC(O5)C(C(C(O)C5COC(C=Cc(c6)ccc(O)c6O)=O)O)O)OC(C(O)C4O)OC(C2=O)=C(Oc(c3)c2c(cc3O)O)c(c1)ccc(O)c1O | + | SMILES C(C4O)(COC(O5)C(C(C(O)C5COC(C=Cc(c6)ccc(O)c6O)=O)O)O)OC(C(O)C4O)OC(C2=O)=C(Oc(c3)c2c(cc3O)O)c(c1)ccc(O)c1O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
56 61 0 0 0 0 0 0 0 0999 V2000
4.2899 1.3375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5754 1.7500 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5754 2.5750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2899 2.9875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.0044 2.5750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.0044 1.7500 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8610 1.3375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1465 1.7500 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4320 1.3375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4320 0.5125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1465 0.1000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8610 0.5125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7175 1.7500 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0031 1.3375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0031 0.5125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7175 0.1000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1465 -0.5608 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7114 1.7500 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4599 0.0670 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7175 -0.6267 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.5963 2.9167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2899 3.7133 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0031 -1.7616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8087 -1.9407 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1943 -1.2110 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8979 -0.7797 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0923 -0.6005 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7066 -1.3301 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1688 -1.0498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6403 -1.2161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0031 -2.4631 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3145 -2.0962 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5500 -1.7700 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1999 -2.9425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6057 -3.1216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9913 -2.3919 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6949 -1.9606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8893 -1.7814 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5036 -2.5111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0342 -2.2307 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1999 -3.7133 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1115 -3.2771 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3469 -2.9509 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6507 -2.2923 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0627 -1.5788 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6507 -0.8653 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8865 -1.5788 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2985 -2.2923 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1224 -2.2923 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5349 -1.5778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3599 -1.5778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7724 -2.2923 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3599 -3.0068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5349 -3.0068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.5963 -2.2923 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.7718 -0.8643 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
2 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 11 1 0 0 0 0
11 12 1 0 0 0 0
12 7 2 0 0 0 0
9 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 10 1 0 0 0 0
14 18 1 0 0 0 0
11 17 2 0 0 0 0
12 19 1 0 0 0 0
16 20 1 0 0 0 0
5 21 1 0 0 0 0
4 22 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
28 29 1 0 0 0 0
29 30 1 0 0 0 0
23 31 1 0 0 0 0
24 32 1 0 0 0 0
25 33 1 0 0 0 0
34 35 1 1 0 0 0
35 36 1 1 0 0 0
37 36 1 1 0 0 0
37 38 1 0 0 0 0
38 39 1 0 0 0 0
39 34 1 0 0 0 0
39 40 1 0 0 0 0
34 41 1 0 0 0 0
35 42 1 0 0 0 0
36 43 1 0 0 0 0
37 30 1 0 0 0 0
26 19 1 0 0 0 0
44 45 1 0 0 0 0
45 46 2 0 0 0 0
45 47 1 0 0 0 0
47 48 2 0 0 0 0
48 49 1 0 0 0 0
49 50 2 0 0 0 0
50 51 1 0 0 0 0
51 52 2 0 0 0 0
52 53 1 0 0 0 0
53 54 2 0 0 0 0
54 49 1 0 0 0 0
52 55 1 0 0 0 0
51 56 1 0 0 0 0
44 40 1 0 0 0 0
S SKP 8
ID FL5FACGS0104
KNApSAcK_ID C00013876
NAME Madreselvin B;Quercetin 3-(6'''-caffeoylgentiobioside);2-(3,4-Dihydroxyphenyl)-3-[[6-O-[6-O-[(2E)-3-(3,4-dihydroxyphenyl)-1-oxo-2-propenyl]-beta-D-glucopyranosyl]-beta-D-glucopyranosyl]oxy]-5,7-dihydroxy-4H-1-benzopyran-4-one
CAS_RN 188682-90-8
FORMULA C36H36O20
EXACTMASS 788.179993592
AVERAGEMASS 788.65904
SMILES C(C4O)(COC(O5)C(C(C(O)C5COC(C=Cc(c6)ccc(O)c6O)=O)O)O)OC(C(O)C4O)OC(C2=O)=C(Oc(c3)c2c(cc3O)O)c(c1)ccc(O)c1O
M END