Mol:FL5FACGS0060
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 53 58 0 0 0 0 0 0 0 0999 V2000 | + | 53 58 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.7697 0.6968 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7697 0.6968 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7697 -0.1308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7697 -0.1308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0538 -0.5447 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0538 -0.5447 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3392 -0.1308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3392 -0.1308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3392 0.6968 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3392 0.6968 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0538 1.1093 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0538 1.1093 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3767 -0.5447 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3767 -0.5447 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0912 -0.1308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0912 -0.1308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0912 0.6968 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0912 0.6968 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3767 1.1093 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3767 1.1093 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8086 1.1093 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8086 1.1093 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5259 0.6968 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5259 0.6968 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2419 1.1093 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2419 1.1093 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2419 1.9370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2419 1.9370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5259 2.3494 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5259 2.3494 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8086 1.9370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8086 1.9370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6914 -0.4259 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6914 -0.4259 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9592 2.3494 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9592 2.3494 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0538 -1.3752 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0538 -1.3752 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3767 -1.3752 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3767 -1.3752 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9138 2.3583 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9138 2.3583 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6748 2.5437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6748 2.5437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2606 1.7962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2606 1.7962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3212 2.5712 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3212 2.5712 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5038 2.3214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5038 2.3214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6245 1.9621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6245 1.9621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3973 1.0445 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3973 1.0445 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9694 2.8939 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9694 2.8939 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0780 2.0450 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0780 2.0450 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9101 1.8237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9101 1.8237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6734 3.1934 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6734 3.1934 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9378 -0.1915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9378 -0.1915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3200 -0.8813 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3200 -0.8813 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4184 -0.1661 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4184 -0.1661 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1727 -0.3976 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1727 -0.3976 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8507 -0.7846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8507 -0.7846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1723 -1.2812 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1723 -1.2812 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9138 -0.6800 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9138 -0.6800 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5657 -0.6517 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5657 -0.6517 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7987 -0.8559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7987 -0.8559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3621 -2.2078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3621 -2.2078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1445 -2.9657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1445 -2.9657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3059 -1.1154 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3059 -1.1154 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6754 -1.8124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6754 -1.8124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7298 -3.0099 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7298 -3.0099 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1499 -2.5496 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1499 -2.5496 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8742 -3.1934 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8742 -3.1934 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7735 -2.2700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7735 -2.2700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0869 -1.8721 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0869 -1.8721 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8479 -2.6228 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8479 -2.6228 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5259 2.9867 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5259 2.9867 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7619 -0.2139 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7619 -0.2139 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8568 0.3227 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8568 0.3227 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 1 6 1 0 0 0 0 | + | 1 6 1 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 5 10 1 0 0 0 0 | + | 5 10 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 11 16 1 0 0 0 0 | + | 11 16 1 0 0 0 0 |
| − | 8 17 1 0 0 0 0 | + | 8 17 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 7 20 2 0 0 0 0 | + | 7 20 2 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 29 1 1 0 0 0 | + | 23 29 1 1 0 0 0 |
| − | 24 30 1 1 0 0 0 | + | 24 30 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 22 25 1 0 0 0 0 | + | 22 25 1 0 0 0 0 |
| − | 21 24 1 0 0 0 0 | + | 21 24 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 23 27 1 0 0 0 0 | + | 23 27 1 0 0 0 0 |
| − | 25 28 1 0 0 0 0 | + | 25 28 1 0 0 0 0 |
| − | 29 30 1 1 0 0 0 | + | 29 30 1 1 0 0 0 |
| − | 1 27 1 0 0 0 0 | + | 1 27 1 0 0 0 0 |
| − | 22 31 1 0 0 0 0 | + | 22 31 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 39 1 1 0 0 0 | + | 33 39 1 1 0 0 0 |
| − | 34 40 1 1 0 0 0 | + | 34 40 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 32 35 1 0 0 0 0 | + | 32 35 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 37 39 1 0 0 0 0 | + | 37 39 1 0 0 0 0 |
| − | 33 38 1 0 0 0 0 | + | 33 38 1 0 0 0 0 |
| − | 39 40 1 1 0 0 0 | + | 39 40 1 1 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 48 1 1 0 0 0 | + | 42 48 1 1 0 0 0 |
| − | 43 49 1 1 0 0 0 | + | 43 49 1 1 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 41 44 1 0 0 0 0 | + | 41 44 1 0 0 0 0 |
| − | 45 48 1 0 0 0 0 | + | 45 48 1 0 0 0 0 |
| − | 44 46 1 0 0 0 0 | + | 44 46 1 0 0 0 0 |
| − | 42 47 1 0 0 0 0 | + | 42 47 1 0 0 0 0 |
| − | 48 49 1 1 0 0 0 | + | 48 49 1 1 0 0 0 |
| − | 50 41 1 0 0 0 0 | + | 50 41 1 0 0 0 0 |
| − | 36 43 1 0 0 0 0 | + | 36 43 1 0 0 0 0 |
| − | 17 34 1 0 0 0 0 | + | 17 34 1 0 0 0 0 |
| − | 15 51 1 0 0 0 0 | + | 15 51 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 32 52 1 0 0 0 0 | + | 32 52 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 52 53 | + | M SAL 1 2 52 53 |
| − | M SBL 1 1 58 | + | M SBL 1 1 58 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 58 -0.8241 0.0224 | + | M SBV 1 58 -0.8241 0.0224 |
| − | S SKP 5 | + | S SKP 5 |
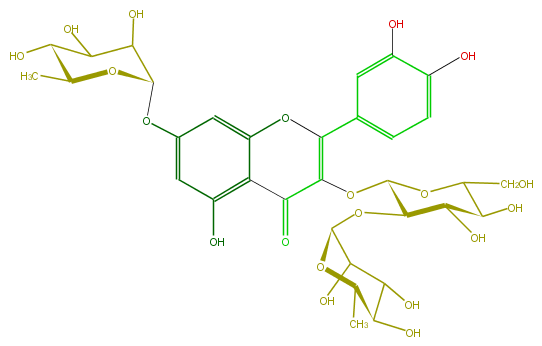
| − | ID FL5FACGS0060 | + | ID FL5FACGS0060 |
| − | FORMULA C33H40O20 | + | FORMULA C33H40O20 |
| − | EXACTMASS 756.21129372 | + | EXACTMASS 756.21129372 |
| − | AVERAGEMASS 756.6587 | + | AVERAGEMASS 756.6587 |
| − | SMILES OC(C6O)C(C(OC(CO)6)OC(=C(c(c5)ccc(c(O)5)O)4)C(=O)c(c2O4)c(cc(OC(O3)C(O)C(O)C(C3C)O)c2)O)OC(C(O)1)OC(C(O)C1O)C | + | SMILES OC(C6O)C(C(OC(CO)6)OC(=C(c(c5)ccc(c(O)5)O)4)C(=O)c(c2O4)c(cc(OC(O3)C(O)C(O)C(C3C)O)c2)O)OC(C(O)1)OC(C(O)C1O)C |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
53 58 0 0 0 0 0 0 0 0999 V2000
-1.7697 0.6968 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7697 -0.1308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0538 -0.5447 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3392 -0.1308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3392 0.6968 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0538 1.1093 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3767 -0.5447 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0912 -0.1308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0912 0.6968 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3767 1.1093 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8086 1.1093 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5259 0.6968 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2419 1.1093 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2419 1.9370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5259 2.3494 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8086 1.9370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6914 -0.4259 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9592 2.3494 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0538 -1.3752 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3767 -1.3752 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9138 2.3583 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6748 2.5437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2606 1.7962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3212 2.5712 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5038 2.3214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6245 1.9621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3973 1.0445 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9694 2.8939 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0780 2.0450 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9101 1.8237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6734 3.1934 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9378 -0.1915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3200 -0.8813 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4184 -0.1661 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1727 -0.3976 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8507 -0.7846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1723 -1.2812 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9138 -0.6800 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5657 -0.6517 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7987 -0.8559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3621 -2.2078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1445 -2.9657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3059 -1.1154 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6754 -1.8124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7298 -3.0099 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1499 -2.5496 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8742 -3.1934 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7735 -2.2700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0869 -1.8721 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8479 -2.6228 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5259 2.9867 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.7619 -0.2139 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8568 0.3227 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
1 6 1 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
5 10 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
11 16 1 0 0 0 0
8 17 1 0 0 0 0
14 18 1 0 0 0 0
3 19 1 0 0 0 0
7 20 2 0 0 0 0
22 23 1 0 0 0 0
23 29 1 1 0 0 0
24 30 1 1 0 0 0
24 25 1 0 0 0 0
22 25 1 0 0 0 0
21 24 1 0 0 0 0
26 30 1 0 0 0 0
23 27 1 0 0 0 0
25 28 1 0 0 0 0
29 30 1 1 0 0 0
1 27 1 0 0 0 0
22 31 1 0 0 0 0
32 33 1 0 0 0 0
33 39 1 1 0 0 0
34 40 1 1 0 0 0
34 35 1 0 0 0 0
32 35 1 0 0 0 0
36 40 1 0 0 0 0
37 39 1 0 0 0 0
33 38 1 0 0 0 0
39 40 1 1 0 0 0
41 42 1 0 0 0 0
42 48 1 1 0 0 0
43 49 1 1 0 0 0
43 44 1 0 0 0 0
41 44 1 0 0 0 0
45 48 1 0 0 0 0
44 46 1 0 0 0 0
42 47 1 0 0 0 0
48 49 1 1 0 0 0
50 41 1 0 0 0 0
36 43 1 0 0 0 0
17 34 1 0 0 0 0
15 51 1 0 0 0 0
52 53 1 0 0 0 0
32 52 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 52 53
M SBL 1 1 58
M SMT 1 CH2OH
M SBV 1 58 -0.8241 0.0224
S SKP 5
ID FL5FACGS0060
FORMULA C33H40O20
EXACTMASS 756.21129372
AVERAGEMASS 756.6587
SMILES OC(C6O)C(C(OC(CO)6)OC(=C(c(c5)ccc(c(O)5)O)4)C(=O)c(c2O4)c(cc(OC(O3)C(O)C(O)C(C3C)O)c2)O)OC(C(O)1)OC(C(O)C1O)C
M END