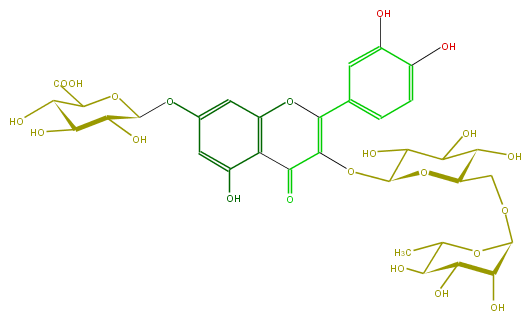
Mol:FL5FACGL0043
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 55 60 0 0 0 0 0 0 0 0999 V2000 | + | 55 60 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.4779 0.9778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4779 0.9778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4779 0.1569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4779 0.1569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7669 -0.2536 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7669 -0.2536 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0559 0.1569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0559 0.1569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0559 0.9778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0559 0.9778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7669 1.3884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7669 1.3884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6551 -0.2536 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6551 -0.2536 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3662 0.1569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3662 0.1569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3662 0.9778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3662 0.9778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6551 1.3884 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6551 1.3884 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6551 -0.9377 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6551 -0.9377 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0769 1.3882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0769 1.3882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8016 0.9698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8016 0.9698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5263 1.3882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5263 1.3882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5263 2.2250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5263 2.2250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8016 2.6434 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8016 2.6434 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0769 2.2250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0769 2.2250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1887 1.3882 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1887 1.3882 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0999 -0.2764 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0999 -0.2764 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7669 -0.9161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7669 -0.9161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2605 -1.9701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2605 -1.9701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8315 -2.7132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8315 -2.7132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6565 -2.4774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6565 -2.4774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4867 -2.7132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4867 -2.7132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.9158 -1.9701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.9158 -1.9701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0907 -2.2058 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0907 -2.2058 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4636 -2.1836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4636 -2.1836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4360 0.2152 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4360 0.2152 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0069 -0.5279 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0069 -0.5279 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8319 -0.2921 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8319 -0.2921 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6621 -0.5279 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6621 -0.5279 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0913 0.2152 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0913 0.2152 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2661 -0.0205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2661 -0.0205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6198 0.1654 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6198 0.1654 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8341 0.6284 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8341 0.6284 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.8984 0.0901 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.8984 0.0901 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4219 -0.3809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4219 -0.3809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.7406 -1.1895 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.7406 -1.1895 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2963 -2.5698 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2963 -2.5698 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1606 -3.1617 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1606 -3.1617 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4880 -3.4799 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4880 -3.4799 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3365 2.6928 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3365 2.6928 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8016 3.4799 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8016 3.4799 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9332 1.3778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9332 1.3778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4149 0.6938 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4149 0.6938 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6687 0.9841 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6687 0.9841 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8910 0.9755 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8910 0.9755 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4719 1.5151 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4719 1.5151 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2341 1.2415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2341 1.2415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.7191 0.9241 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.7191 0.9241 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2228 0.6546 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2228 0.6546 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9923 0.4918 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9923 0.4918 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8614 1.8217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8614 1.8217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.9158 1.8217 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.9158 1.8217 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3342 2.7348 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3342 2.7348 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 8 19 1 0 0 0 0 | + | 8 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 30 1 1 0 0 0 | + | 29 30 1 1 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 28 1 0 0 0 0 | + | 33 28 1 0 0 0 0 |
| − | 28 34 1 0 0 0 0 | + | 28 34 1 0 0 0 0 |
| − | 33 35 1 0 0 0 0 | + | 33 35 1 0 0 0 0 |
| − | 32 36 1 0 0 0 0 | + | 32 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 25 38 1 0 0 0 0 | + | 25 38 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 22 39 1 0 0 0 0 | + | 22 39 1 0 0 0 0 |
| − | 23 40 1 0 0 0 0 | + | 23 40 1 0 0 0 0 |
| − | 24 41 1 0 0 0 0 | + | 24 41 1 0 0 0 0 |
| − | 29 19 1 0 0 0 0 | + | 29 19 1 0 0 0 0 |
| − | 42 15 1 0 0 0 0 | + | 42 15 1 0 0 0 0 |
| − | 16 43 1 0 0 0 0 | + | 16 43 1 0 0 0 0 |
| − | 44 45 1 1 0 0 0 | + | 44 45 1 1 0 0 0 |
| − | 45 46 1 1 0 0 0 | + | 45 46 1 1 0 0 0 |
| − | 47 46 1 1 0 0 0 | + | 47 46 1 1 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 49 44 1 0 0 0 0 | + | 49 44 1 0 0 0 0 |
| − | 44 50 1 0 0 0 0 | + | 44 50 1 0 0 0 0 |
| − | 45 51 1 0 0 0 0 | + | 45 51 1 0 0 0 0 |
| − | 46 52 1 0 0 0 0 | + | 46 52 1 0 0 0 0 |
| − | 47 18 1 0 0 0 0 | + | 47 18 1 0 0 0 0 |
| − | 53 54 2 0 0 0 0 | + | 53 54 2 0 0 0 0 |
| − | 53 55 1 0 0 0 0 | + | 53 55 1 0 0 0 0 |
| − | 49 53 1 0 0 0 0 | + | 49 53 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 3 53 54 55 | + | M SAL 1 3 53 54 55 |
| − | M SBL 1 1 60 | + | M SBL 1 1 60 |
| − | M SMT 1 ^COOH | + | M SMT 1 ^COOH |
| − | M SBV 1 60 0.6272 -0.5803 | + | M SBV 1 60 0.6272 -0.5803 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FACGL0043 | + | ID FL5FACGL0043 |
| − | FORMULA C33H38O22 | + | FORMULA C33H38O22 |
| − | EXACTMASS 786.1854728999999 | + | EXACTMASS 786.1854728999999 |
| − | AVERAGEMASS 786.64162 | + | AVERAGEMASS 786.64162 |
| − | SMILES O(C(C6O)OC(C(O)=O)C(O)C(O)6)c(c5)cc(c(c(O)5)2)OC(=C(OC(C(O)3)OC(COC(C4O)OC(C(C4O)O)C)C(O)C3O)C2=O)c(c1)ccc(O)c(O)1 | + | SMILES O(C(C6O)OC(C(O)=O)C(O)C(O)6)c(c5)cc(c(c(O)5)2)OC(=C(OC(C(O)3)OC(COC(C4O)OC(C(C4O)O)C)C(O)C3O)C2=O)c(c1)ccc(O)c(O)1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
55 60 0 0 0 0 0 0 0 0999 V2000
-1.4779 0.9778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4779 0.1569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7669 -0.2536 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0559 0.1569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0559 0.9778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7669 1.3884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6551 -0.2536 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3662 0.1569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3662 0.9778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6551 1.3884 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6551 -0.9377 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0769 1.3882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8016 0.9698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5263 1.3882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5263 2.2250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8016 2.6434 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0769 2.2250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1887 1.3882 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0999 -0.2764 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7669 -0.9161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2605 -1.9701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8315 -2.7132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6565 -2.4774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.4867 -2.7132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.9158 -1.9701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.0907 -2.2058 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4636 -2.1836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4360 0.2152 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0069 -0.5279 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8319 -0.2921 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6621 -0.5279 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.0913 0.2152 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2661 -0.0205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6198 0.1654 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.8341 0.6284 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.8984 0.0901 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.4219 -0.3809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.7406 -1.1895 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2963 -2.5698 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1606 -3.1617 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.4880 -3.4799 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.3365 2.6928 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8016 3.4799 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9332 1.3778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4149 0.6938 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6687 0.9841 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8910 0.9755 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4719 1.5151 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2341 1.2415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.7191 0.9241 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.2228 0.6546 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9923 0.4918 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8614 1.8217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.9158 1.8217 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3342 2.7348 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
8 19 1 0 0 0 0
3 20 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
28 29 1 0 0 0 0
29 30 1 1 0 0 0
30 31 1 1 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 28 1 0 0 0 0
28 34 1 0 0 0 0
33 35 1 0 0 0 0
32 36 1 0 0 0 0
31 37 1 0 0 0 0
25 38 1 0 0 0 0
37 38 1 0 0 0 0
22 39 1 0 0 0 0
23 40 1 0 0 0 0
24 41 1 0 0 0 0
29 19 1 0 0 0 0
42 15 1 0 0 0 0
16 43 1 0 0 0 0
44 45 1 1 0 0 0
45 46 1 1 0 0 0
47 46 1 1 0 0 0
47 48 1 0 0 0 0
48 49 1 0 0 0 0
49 44 1 0 0 0 0
44 50 1 0 0 0 0
45 51 1 0 0 0 0
46 52 1 0 0 0 0
47 18 1 0 0 0 0
53 54 2 0 0 0 0
53 55 1 0 0 0 0
49 53 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 3 53 54 55
M SBL 1 1 60
M SMT 1 ^COOH
M SBV 1 60 0.6272 -0.5803
S SKP 5
ID FL5FACGL0043
FORMULA C33H38O22
EXACTMASS 786.1854728999999
AVERAGEMASS 786.64162
SMILES O(C(C6O)OC(C(O)=O)C(O)C(O)6)c(c5)cc(c(c(O)5)2)OC(=C(OC(C(O)3)OC(COC(C4O)OC(C(C4O)O)C)C(O)C3O)C2=O)c(c1)ccc(O)c(O)1
M END