Mol:FL5FACGA0021
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 54 59 0 0 0 0 0 0 0 0999 V2000 | + | 54 59 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.7298 1.2402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7298 1.2402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7298 0.5979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7298 0.5979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1735 0.2767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1735 0.2767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3828 0.5979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3828 0.5979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3828 1.2402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3828 1.2402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1735 1.5614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1735 1.5614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9391 0.2767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9391 0.2767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4954 0.5979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4954 0.5979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4954 1.2402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4954 1.2402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9391 1.5614 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9391 1.5614 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9391 -0.2241 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9391 -0.2241 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0515 1.5613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0515 1.5613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6184 1.2340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6184 1.2340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1854 1.5613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1854 1.5613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1854 2.2160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1854 2.2160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6184 2.5433 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6184 2.5433 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0515 2.2160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0515 2.2160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1735 -0.3654 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1735 -0.3654 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2859 1.5613 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2859 1.5613 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7522 2.5432 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7522 2.5432 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8895 0.2090 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8895 0.2090 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6781 -0.8182 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.6781 -0.8182 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.0067 -0.4305 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.0067 -0.4305 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.2198 -1.1760 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2198 -1.1760 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0067 -1.9260 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.0067 -1.9260 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.6781 -2.3137 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.6781 -2.3137 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.4651 -1.5682 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.4651 -1.5682 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.4852 -0.0982 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4852 -0.0982 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0767 -1.6216 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0767 -1.6216 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8693 -2.8391 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8693 -2.8391 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5553 -0.9201 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 3.5553 -0.9201 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 4.2807 -0.9201 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 4.2807 -0.9201 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.7593 -0.4157 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 3.7593 -0.4157 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 3.5811 0.2917 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5811 0.2917 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8556 0.2917 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.8556 0.2917 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.3770 -0.2127 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.3770 -0.2127 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 4.0442 0.1725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0442 0.1725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9961 -1.3609 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9961 -1.3609 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4323 -1.4858 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4323 -1.4858 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4755 -0.4157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4755 -0.4157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6184 3.1978 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6184 3.1978 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7617 1.2196 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -3.7617 1.2196 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -3.2461 0.5390 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.2461 0.5390 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.5036 0.8277 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.5036 0.8277 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.7871 0.8355 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.7871 0.8355 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.3078 1.3562 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3078 1.3562 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9573 1.0134 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -2.9573 1.0134 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -4.4755 0.8075 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4755 0.8075 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0499 0.4999 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0499 0.4999 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0782 0.1135 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0782 0.1135 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0579 1.8710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0579 1.8710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9763 2.2667 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9763 2.2667 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3621 -2.4583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3621 -2.4583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8319 -3.0903 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8319 -3.0903 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 8 21 1 0 0 0 0 | + | 8 21 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 23 21 1 0 0 0 0 | + | 23 21 1 0 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 1 0 0 0 | + | 35 36 1 1 0 0 0 |
| − | 36 31 1 1 0 0 0 | + | 36 31 1 1 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 31 38 1 0 0 0 0 | + | 31 38 1 0 0 0 0 |
| − | 32 39 1 0 0 0 0 | + | 32 39 1 0 0 0 0 |
| − | 33 40 1 0 0 0 0 | + | 33 40 1 0 0 0 0 |
| − | 35 28 1 0 0 0 0 | + | 35 28 1 0 0 0 0 |
| − | 16 41 1 0 0 0 0 | + | 16 41 1 0 0 0 0 |
| − | 42 43 1 1 0 0 0 | + | 42 43 1 1 0 0 0 |
| − | 43 44 1 1 0 0 0 | + | 43 44 1 1 0 0 0 |
| − | 45 44 1 1 0 0 0 | + | 45 44 1 1 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 42 1 0 0 0 0 | + | 47 42 1 0 0 0 0 |
| − | 42 48 1 0 0 0 0 | + | 42 48 1 0 0 0 0 |
| − | 43 49 1 0 0 0 0 | + | 43 49 1 0 0 0 0 |
| − | 44 50 1 0 0 0 0 | + | 44 50 1 0 0 0 0 |
| − | 45 19 1 0 0 0 0 | + | 45 19 1 0 0 0 0 |
| − | 47 51 1 0 0 0 0 | + | 47 51 1 0 0 0 0 |
| − | 51 52 1 0 0 0 0 | + | 51 52 1 0 0 0 0 |
| − | 25 53 1 0 0 0 0 | + | 25 53 1 0 0 0 0 |
| − | 53 54 1 0 0 0 0 | + | 53 54 1 0 0 0 0 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 53 54 | + | M SAL 2 2 53 54 |
| − | M SBL 2 1 58 | + | M SBL 2 1 58 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 58 2.4808 -2.6145 | + | M SVB 2 58 2.4808 -2.6145 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 51 52 | + | M SAL 1 2 51 52 |
| − | M SBL 1 1 56 | + | M SBL 1 1 56 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SVB 1 56 -3.0579 1.871 | + | M SVB 1 56 -3.0579 1.871 |
| − | S SKP 8 | + | S SKP 8 |
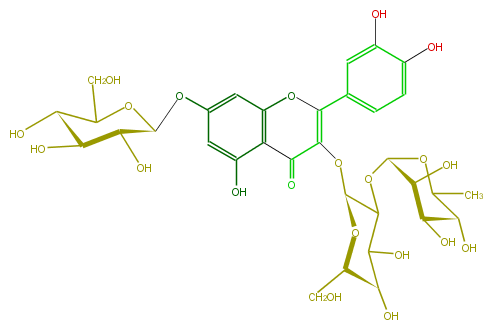
| − | ID FL5FACGA0021 | + | ID FL5FACGA0021 |
| − | KNApSAcK_ID C00005459 | + | KNApSAcK_ID C00005459 |
| − | NAME Quercetin 3-rhamnosyl-(1->2)-galactoside-7-glucoside | + | NAME Quercetin 3-rhamnosyl-(1->2)-galactoside-7-glucoside |
| − | CAS_RN 128988-57-8 | + | CAS_RN 128988-57-8 |
| − | FORMULA C33H40O21 | + | FORMULA C33H40O21 |
| − | EXACTMASS 772.206208342 | + | EXACTMASS 772.206208342 |
| − | AVERAGEMASS 772.6581 | + | AVERAGEMASS 772.6581 |
| − | SMILES CC(O1)[C@@H](O)[C@H]([C@@H]([C@H]1OC([C@@H]2OC(C(=O)5)=C(c(c6)cc(c(c6)O)O)Oc(c53)cc(O[C@@H]([C@@H](O)4)OC(CO)[C@H](O)[C@@H]4O)cc3O)C(O)[C@H]([C@@H](CO)O2)O)O)O | + | SMILES CC(O1)[C@@H](O)[C@H]([C@@H]([C@H]1OC([C@@H]2OC(C(=O)5)=C(c(c6)cc(c(c6)O)O)Oc(c53)cc(O[C@@H]([C@@H](O)4)OC(CO)[C@H](O)[C@@H]4O)cc3O)C(O)[C@H]([C@@H](CO)O2)O)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
54 59 0 0 0 0 0 0 0 0999 V2000
-0.7298 1.2402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7298 0.5979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1735 0.2767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3828 0.5979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3828 1.2402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1735 1.5614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9391 0.2767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4954 0.5979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4954 1.2402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9391 1.5614 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9391 -0.2241 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0515 1.5613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6184 1.2340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1854 1.5613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1854 2.2160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6184 2.5433 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0515 2.2160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1735 -0.3654 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2859 1.5613 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7522 2.5432 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8895 0.2090 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6781 -0.8182 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.0067 -0.4305 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.2198 -1.1760 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0067 -1.9260 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.6781 -2.3137 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.4651 -1.5682 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.4852 -0.0982 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0767 -1.6216 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8693 -2.8391 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5553 -0.9201 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
4.2807 -0.9201 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.7593 -0.4157 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
3.5811 0.2917 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8556 0.2917 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.3770 -0.2127 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
4.0442 0.1725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9961 -1.3609 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.4323 -1.4858 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.4755 -0.4157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6184 3.1978 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7617 1.2196 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-3.2461 0.5390 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.5036 0.8277 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.7871 0.8355 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.3078 1.3562 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9573 1.0134 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-4.4755 0.8075 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0499 0.4999 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0782 0.1135 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0579 1.8710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9763 2.2667 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3621 -2.4583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8319 -3.0903 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
15 20 1 0 0 0 0
8 21 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
23 21 1 0 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 36 1 1 0 0 0
36 31 1 1 0 0 0
36 37 1 0 0 0 0
31 38 1 0 0 0 0
32 39 1 0 0 0 0
33 40 1 0 0 0 0
35 28 1 0 0 0 0
16 41 1 0 0 0 0
42 43 1 1 0 0 0
43 44 1 1 0 0 0
45 44 1 1 0 0 0
45 46 1 0 0 0 0
46 47 1 0 0 0 0
47 42 1 0 0 0 0
42 48 1 0 0 0 0
43 49 1 0 0 0 0
44 50 1 0 0 0 0
45 19 1 0 0 0 0
47 51 1 0 0 0 0
51 52 1 0 0 0 0
25 53 1 0 0 0 0
53 54 1 0 0 0 0
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 53 54
M SBL 2 1 58
M SMT 2 CH2OH
M SVB 2 58 2.4808 -2.6145
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 51 52
M SBL 1 1 56
M SMT 1 CH2OH
M SVB 1 56 -3.0579 1.871
S SKP 8
ID FL5FACGA0021
KNApSAcK_ID C00005459
NAME Quercetin 3-rhamnosyl-(1->2)-galactoside-7-glucoside
CAS_RN 128988-57-8
FORMULA C33H40O21
EXACTMASS 772.206208342
AVERAGEMASS 772.6581
SMILES CC(O1)[C@@H](O)[C@H]([C@@H]([C@H]1OC([C@@H]2OC(C(=O)5)=C(c(c6)cc(c(c6)O)O)Oc(c53)cc(O[C@@H]([C@@H](O)4)OC(CO)[C@H](O)[C@@H]4O)cc3O)C(O)[C@H]([C@@H](CO)O2)O)O)O
M END